miRNA Alterations Elicit Pathways Involved in Memory Decline and Synaptic Function in the Hippocampus of Aged Tg4-42 Mice
- PMID: 33013313
- PMCID: PMC7511553
- DOI: 10.3389/fnins.2020.580524
miRNA Alterations Elicit Pathways Involved in Memory Decline and Synaptic Function in the Hippocampus of Aged Tg4-42 Mice
Abstract
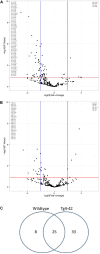
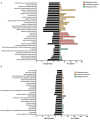
The transcriptome of non-coding RNA (ncRNA) species is increasingly focused in Alzheimer's disease (AD) research. NcRNAs comprise, among others, transfer RNAs, long non-coding RNAs and microRNAs (miRs), each with their own specific biological function. We used smallRNASeq to assess miR expression in the hippocampus of young (3 month old) and aged (8 month old) Tg4-42 mice, a model system for sporadic AD, as well as age-matched wildtype controls. Tg4-42 mice express N-truncated Aβ4-42, develop age-related neuron loss, reduced neurogenesis and behavioral deficits. Our results do not only confirm known miR-AD associations in Tg4-42 mice, but more importantly pinpoint 22 additional miRs associated to the disease. Twenty-five miRs were differentially expressed in both aged Tg4-42 and aged wildtype mice while eight miRs were differentially expressed only in aged wildtype mice, and 33 only in aged Tg4-42 mice. No significant alteration in the miRNome was detected in young mice, which indicates that the changes observed in aged mice are down-stream effects of Aβ-induced pathology in the Tg4-42 mouse model for AD. Targets of those miRs were predicted using miRWalk. For miRs that were differentially expressed only in the Tg4-42 model, 128 targets could be identified, whereas 18 genes were targeted by miRs only differentially expressed in wildtype mice and 85 genes were targeted by miRs differentially expressed in both mouse models. Genes targeted by differentially expressed miRs in the Tg4-42 model were enriched for negative regulation of long-term synaptic potentiation, learning or memory, regulation of trans-synaptic signaling and modulation of chemical synaptic transmission obtained. This untargeted miR sequencing approach supports previous reports on the Tg4-42 mice as a valuable model for AD. Furthermore, it revealed miRs involved in AD, which can serve as biomarkers or therapeutic targets.
Keywords: Alzheimer; NGS; Tg4-42; miRNA transcriptome; miRNA-Seq; transgenic mouse model.
Copyright © 2020 Bouter, Kacprowski, Rößler, Jensen, Kuss and Bayer.
Figures

Similar articles
-
Small RNA Sequencing in the Tg4-42 Mouse Model Suggests the Involvement of snoRNAs in the Etiology of Alzheimer's Disease.J Alzheimers Dis. 2022;87(4):1671-1681. doi: 10.3233/JAD-220110. J Alzheimers Dis. 2022. PMID: 35527555
-
Synaptic Alterations in Mouse Models for Alzheimer Disease-A Special Focus on N-Truncated Abeta 4-42.Molecules. 2018 Mar 21;23(4):718. doi: 10.3390/molecules23040718. Molecules. 2018. PMID: 29561816 Free PMC article. Review.
-
Deciphering the molecular profile of plaques, memory decline and neuron loss in two mouse models for Alzheimer's disease by deep sequencing.Front Aging Neurosci. 2014 Apr 16;6:75. doi: 10.3389/fnagi.2014.00075. eCollection 2014. Front Aging Neurosci. 2014. PMID: 24795628 Free PMC article.
-
Metabolic, Phenotypic, and Neuropathological Characterization of the Tg4-42 Mouse Model for Alzheimer's Disease.J Alzheimers Dis. 2021;80(3):1151-1168. doi: 10.3233/JAD-201204. J Alzheimers Dis. 2021. PMID: 33646155 Free PMC article.
-
Synaptic Dysfunction in Alzheimer's Disease: Aβ, Tau, and Epigenetic Alterations.Mol Neurobiol. 2018 Apr;55(4):3021-3032. doi: 10.1007/s12035-017-0533-3. Epub 2017 Apr 29. Mol Neurobiol. 2018. PMID: 28456942 Review.
Cited by
-
MicroRNA Dysregulation in the Hippocampus of Rats with Noise-Induced Hearing Loss.Oxid Med Cell Longev. 2021 Sep 6;2021:1377195. doi: 10.1155/2021/1377195. eCollection 2021. Oxid Med Cell Longev. 2021. PMID: 34527169 Free PMC article.
-
N-Truncated Aβ Starting at Position Four-Biochemical Features, Preclinical Models, and Potential as Drug Target in Alzheimer's Disease.Front Aging Neurosci. 2021 Aug 20;13:710579. doi: 10.3389/fnagi.2021.710579. eCollection 2021. Front Aging Neurosci. 2021. PMID: 34489680 Free PMC article.
-
Fission Impossible: Stabilized miRNA-Based Analogs in Neurodegenerative Disease.Front Neurosci. 2022 May 3;16:875957. doi: 10.3389/fnins.2022.875957. eCollection 2022. Front Neurosci. 2022. PMID: 35592255 Free PMC article. No abstract available.
References
-
- Bartel D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116 281–297. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

