Age-Dependent Assessment of Genes Involved in Cellular Senescence, Telomere, and Mitochondrial Pathways in Human Lung Tissue of Smokers, COPD, and IPF: Associations With SARS-CoV-2 COVID-19 ACE2-TMPRSS2-Furin-DPP4 Axis
- PMID: 33013423
- PMCID: PMC7510459
- DOI: 10.3389/fphar.2020.584637
Age-Dependent Assessment of Genes Involved in Cellular Senescence, Telomere, and Mitochondrial Pathways in Human Lung Tissue of Smokers, COPD, and IPF: Associations With SARS-CoV-2 COVID-19 ACE2-TMPRSS2-Furin-DPP4 Axis
Abstract
Background: Aging is one of the key contributing factors for chronic obstructive pulmonary diseases (COPD) and other chronic inflammatory lung diseases. Here, we determined how aging contributes to the altered gene expression related to mitochondrial function, cellular senescence, and telomeric length processes that play an important role in the progression of COPD and idiopathic pulmonary fibrosis (IPF).
Methods: Total RNA from the human lung tissues of non-smokers, smokers, and patients with COPD and IPF were processed and analyzed using a Nanostring platform based on their ages (younger: <55 years and older: >55 years).
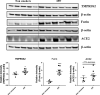
Results: Several genes were differentially expressed in younger and older smokers, and patients with COPD and IPF compared to non-smokers which were part of the mitochondrial biogenesis/function (HSPD1, FEN1, COX18, COX10, UCP2 & 3), cellular senescence (PCNA, PTEN, KLOTHO, CDKN1C, TNKS2, NFATC1 & 2, GADD45A), and telomere replication/maintenance (PARP1, SIRT6, NBN, TERT, RAD17, SLX4, HAT1) target genes. Interestingly, NOX4 and TNKS2 were increased in the young IPF as compared to the young COPD patients. Genes in the mitochondrial dynamics and quality control mechanisms like FIS1 and RHOT2 were decreased in young IPF compared to their age matched COPD subjects. ERCC1 and GADD45B were higher in young COPD as compared to IPF. Aging plays an important role in various infectious diseases including the SARS-CoV-2 infection. Lung immunoblot analysis of smokers, COPD and IPF subjects revealed increased abundance of proteases and receptor/spike protein like TMPRSS2, furin, and DPP4 in association with a slight increase in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) receptor ACE2 levels.
Conclusions: Overall, these findings suggest that altered transcription of target genes that regulate mitochondrial function, cellular senescence, and telomere attrition in the pathobiology of lung aging in COPD and IPF is associated with alterations in SARS-CoV-2 ACE2-TMPRSS2-Furin-DPP4 axis as pharmacological targets for COVID-19.
Keywords: DNA damage; aging; cellular senescence; chronic obstructive pulmonary diseases; idiopathic pulmonary fibrosis; mitochondria; smokers; telomere.
Copyright © 2020 Maremanda, Sundar, Li and Rahman.
Figures






Update of
-
Age-dependent assessment of genes involved in cellular senescence, telomere and mitochondrial pathways in human lung tissue of smokers, COPD and IPF: Associations with SARS-CoV-2 COVID-19 ACE2-TMPRSS2-Furin-DPP4 axis.medRxiv [Preprint]. 2020 Jun 16:2020.06.14.20129957. doi: 10.1101/2020.06.14.20129957. medRxiv. 2020. Update in: Front Pharmacol. 2020 Sep 09;11:584637. doi: 10.3389/fphar.2020.584637. PMID: 32587985 Free PMC article. Updated. Preprint.
-
Age-dependent assessment of genes involved in cellular senescence, telomere and mitochondrial pathways in human lung tissue of smokers, COPD and IPF: Associations with SARS-CoV-2 COVID-19 ACE2-TMPRSS2-Furin-DPP4 axis.Res Sq [Preprint]. 2020 Jun 15:rs.3.rs-35347. doi: 10.21203/rs.3.rs-35347/v1. Res Sq. 2020. Update in: Front Pharmacol. 2020 Sep 09;11:584637. doi: 10.3389/fphar.2020.584637. PMID: 32702724 Free PMC article. Updated. Preprint.
References
-
- Ahmad T., Sundar I. K., Tormos A. M., Lerner C. A., Gerloff J., Yao H., et al. (2017). Shelterin Telomere Protection Protein 1 Reduction Causes Telomere Attrition and Cellular Senescence via Sirtuin 1 Deacetylase in Chronic Obstructive Pulmonary Disease. Am. J. Respir. Cell Mol. Biol. 56, 38–49. 10.1165/rcmb.2016-0198OC - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

