Bioorthogonal Non-canonical Amino Acid Tagging Combined With Flow Cytometry for Determination of Activity in Aquatic Microorganisms
- PMID: 33013733
- PMCID: PMC7461810
- DOI: 10.3389/fmicb.2020.01929
Bioorthogonal Non-canonical Amino Acid Tagging Combined With Flow Cytometry for Determination of Activity in Aquatic Microorganisms
Abstract
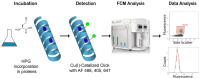
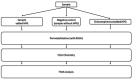
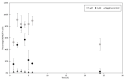
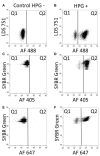
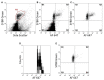
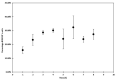
In this study, we have combined bioorthogonal non-canonical amino acid tagging (BONCAT) and flow cytometry (FCM) analysis, and we demonstrate the applicability of the method for marine prokaryotes. Enumeration of active marine bacteria was performed by combining the DNA stain SYBR Green with detection of protein production with BONCAT. After optimization of incubation condition and substrate concentration on monoculture of Escherichia coli, we applied and modified the method to natural marine samples. We found that between 10 and 30% of prokaryotes in natural communities were active. The method is replicable, fast, and allow high sample throughput when using FCM. We conclude that the combination of BONCAT and FCM is an alternative to current methods for quantifying active populations in aquatic environments.
Keywords: bioorthogonal non-canonical amino acid tagging; flow cytometry; marine microbes; protein synthesis; single cell activity.
Copyright © 2020 Lindivat, Larsen, Hess-Erga, Bratbak and Hoell.
Figures



References
-
- Bell R. T. (1993). “Estimating production of heterotrophic bacterioplankton via incorporation of tritiated thymidine” in Handbook of methods in aquatic microbial ecology. 1st Edn. ed. Kemp P. F. (New York: Taylor and Francis; ), 495–504.
-
- Cottrell M. T., Kirchman D. L. (2004). Single-cell analysis of bacterial growth, cell size, and community structure in the Delaware estuary. Aquat. Microb. Ecol. 34, 139–149. 10.3354/ame034139 - DOI
LinkOut - more resources
Full Text Sources

