Microbial Life in the Deep Subsurface Aquifer Illuminated by Metagenomics
- PMID: 33013807
- PMCID: PMC7509429
- DOI: 10.3389/fmicb.2020.572252
Microbial Life in the Deep Subsurface Aquifer Illuminated by Metagenomics
Abstract
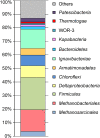
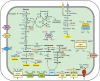
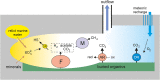
To get insights into microbial diversity and biogeochemical processes in the terrestrial deep subsurface aquifer, we sequenced the metagenome of artesian water collected at a 2.8 km deep oil exploration borehole 5P in Western Siberia, Russia. We obtained 71 metagenome-assembled genomes (MAGs), altogether comprising 93% of the metagenome. Methanogenic archaea accounted for about 20% of the community and mostly belonged to hydrogenotrophic Methanobacteriaceae; acetoclastic and methylotrophic lineages were less abundant. ANME archaea were not found. The most numerous bacteria were the Firmicutes, Ignavibacteriae, Deltaproteobacteria, Chloroflexi, and Armatimonadetes. Most of the community was composed of anaerobic heterotrophs. Only six MAGs belonged to sulfate reducers. These MAGs accounted for 5% of the metagenome and were assigned to the Firmicutes, Deltaproteobacteria, Candidatus Kapabacteria, and Nitrospirae. Organotrophic bacteria carrying cytochrome c oxidase genes and presumably capable of aerobic respiration mostly belonged to the Chloroflexi, Ignavibacteriae, and Armatimonadetes. They accounted for 13% of the community. The first complete closed genomes were obtained for members of the Ignavibacteriae SJA-28 lineage and the candidate phylum Kapabacteria. Metabolic reconstruction of the SJA-28 bacterium, designated Candidatus Tepidiaquacella proteinivora, predicted that it is an anaerobe growing on proteinaceous substrates by fermentation or anaerobic respiration. The Ca. Kapabacteria genome contained both the sulfate reduction pathway and cytochrome c oxidase. Presumably, the availability of buried organic matter of Mesozoic marine sediments, long-term recharge of the aquifer with meteoric waters and its spatial heterogeneity provided the conditions for the development of microbial communities, taxonomically and functionally more diverse than those found in oligotrophic underground ecosystems.
Keywords: candidate phylum; deep subsurface; metagenome; microbial diversity; uncultivable bacteria.
Copyright © 2020 Kadnikov, Mardanov, Beletsky, Karnachuk and Ravin.
Figures

References
-
- Banks D., Frank Y. A., Kadnikov V. V., Watts M., Boyce A., Frengstad B. S. (2014). Hydrochemical Data Report from Sampling of Two Deep Abandoned Hydrocarbon Exploration Wells: Byelii Yar and Parabel’, Tomsk oblast’, Western Siberia, Russian Federation. NGU Report No. 2014.034, Trondheim: Geological Survey of Norway.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

