Excessive Neutrophils and Neutrophil Extracellular Traps in COVID-19
- PMID: 33013872
- PMCID: PMC7461898
- DOI: 10.3389/fimmu.2020.02063
Excessive Neutrophils and Neutrophil Extracellular Traps in COVID-19
Abstract
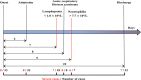
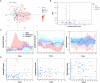
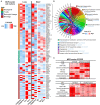
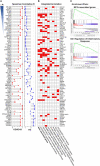
Background: Cases of excessive neutrophil counts in the blood in severe coronavirus disease (COVID-19) patients have drawn significant attention. Neutrophil infiltration was also noted on the pathological findings from autopsies. It is urgent to clarify the pathogenesis of neutrophils leading to severe pneumonia in COVID-19. Methods: A retrospective analysis was performed on 55 COVID-19 patients classified as mild (n = 22), moderate (n = 25), and severe (n = 8) according to the Guidelines released by the National Health Commission of China. Trends relating leukocyte counts and lungs examined by chest CT scan were quantified by Bayesian inference. Transcriptional signatures of host immune cells of four COVID19 patients were analyzed by RNA sequencing of lung specimens and BALF. Results: Neutrophilia occurred in 6 of 8 severe patients at 7-19 days after symptom onset, coinciding with lesion progression. Increasing neutrophil counts paralleled lesion CT values (slope: 0.8 and 0.3-1.2), reflecting neutrophilia-induced lung injury in severe patients. Transcriptome analysis revealed that neutrophil activation was correlated with 17 neutrophil extracellular trap (NET)-associated genes in COVID-19 patients, which was related to innate immunity and interacted with T/NK/B cells, as supported by a protein-protein interaction network analysis. Conclusion: Excessive neutrophils and associated NETs could explain the pathogenesis of lung injury in COVID-19 pneumonia.
Keywords: COVID-19; coronavirus; lymphopenia; neutrophil extracellular trap; neutrophilia; pneumonia.
Copyright © 2020 Wang, Li, Yin, Zhang, Cao, Lin, Huang, Hoffmann, Lu and Qiu.
Figures

References
-
- WHO . Coronavirus Disease (COVID-2019) Situation Reports. Available online at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situatio... (accessed March 30, 2020).
-
- WHO Clinical Management of Severe Acute Respiratory Infection When Novel Coronavirus (nCoV) Infection Is Suspected. Geneva: World Health Organization; (2020). Available online at: https://apps.who.int/iris/handle/10665/330893 (accessed January 28, 2020).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

