Comparative Genomics of Spatholobus suberectus and Insight Into Flavonoid Biosynthesis
- PMID: 33013959
- PMCID: PMC7500164
- DOI: 10.3389/fpls.2020.528108
Comparative Genomics of Spatholobus suberectus and Insight Into Flavonoid Biosynthesis
Abstract
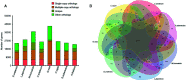
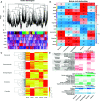
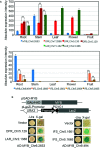
Spatholobus suberectus Dunn (S. suberectus), has been widely used in traditional medicines plant source of the Leguminosae family. Its vine stem of which plays an important role in the prevention and treatment of various diseases because it contains various flavonoids. Comparative genome analysis suggested well-conserved genomic components and genetic collinearity between the genome of S. suberectus and other genera of Leguminosae such as Glycine max. We discovered two whole genome duplications (WGD) events in S. suberectus and G. max lineage underwent a WGD after speciation from S. suberectus. The determination of expansion and contractions of orthologous gene families revealed 1,001 expanded gene families and 3,649 contracted gene families in the S. suberectus lineage. Comparing to the model plants, many novel flavonoid biosynthesis-related genes were predicted in the genome of S. suberectus, and the expression patterns of these genes in the roots are similar to those in the stems [such as the isoflavone synthase (IFS) genes]. The expansion of IFS from a single copy in the Leguminosae ancestor to four copies in S. suberectus, will accelerate the biosynthesis of flavonoids. MYB genes are widely involved in plant flavonoid biosynthesis and the most abundant member of the TF family in S. suberectus. Activated retrotransponson positive regulates the accumulation of flavonoid in S. suberectus by introducing the cis-elements of tissue-specific expressed MYBs. Our study not only provides significant insight into the evolution of specific flavonoid biosynthetic pathways in S. suberectus, but also would facilitate the development of tools for enhancing bioactive productivity by metabolic engineering in microbes or by molecular breeding for alleviating resource shortage of S. suberectus.
Keywords: Spatholobus suberectus Dunn; comparative genome analysis; flavonoid biosynthesis; isoflavone synthase; transcription factors.
Copyright © 2020 Qin, Wei, Cui, Liang, Li, Gu, Yang, Zhou, Li, Xu, Liu, Miao and Zhang.
Figures




References
-
- Dhaubhadel S., McGarvey B. D., Williams R., Gijzen M. (2003). Isoflavonoid biosynthesis and accumulation in developing soybean seeds. Plant Mol. Biol. 53, 733–743. 10.1023/B:PLAN.0000023666.30358.ae - DOI - PubMed
Associated data
LinkOut - more resources
Full Text Sources
Miscellaneous

