Comparative Plastid Genomics of Non-Photosynthetic Chrysophytes: Genome Reduction and Compaction
- PMID: 33013997
- PMCID: PMC7511666
- DOI: 10.3389/fpls.2020.572703
Comparative Plastid Genomics of Non-Photosynthetic Chrysophytes: Genome Reduction and Compaction
Abstract
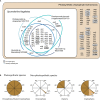
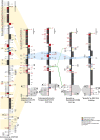
Spumella-like heterotrophic chrysophytes are important eukaryotic microorganisms that feed on bacteria in aquatic and soil environments. They are characterized by their lack of pigmentation, naked cell surface, and extremely small size. Although Spumella-like chrysophytes have lost their photosynthetic ability, they still possess a leucoplast and retain a plastid genome. We have sequenced the plastid genomes of three non-photosynthetic chrysophytes, Spumella sp. Baeckdong012018B8, Pedospumella sp. Jangsampo120217C5 and Poteriospumella lacustris Yongseonkyo072317C3, and compared them to the previously sequenced plastid genome of "Spumella" sp. NIES-1846 and photosynthetic chrysophytes. We found the plastid genomes of Spumella-like flagellates to be generally conserved with respect to genome structure and housekeeping gene content. We nevertheless also observed lineage-specific gene rearrangements and duplication of partial gene fragments at the boundary of the inverted repeat and single copy regions. Most gene losses correspond to genes for proteins involved in photosynthesis and carbon fixation, except in the case of petF. The newly sequenced plastid genomes range from ~55.7 kbp to ~62.9 kbp in size and share a core set of 45 protein-coding genes, 3 rRNAs, and 32 to 34 tRNAs. Our results provide insight into the evolutionary history of organelle genomes via genome reduction and gene loss related to loss of photosynthesis in chrysophyte evolution.
Keywords: chrysophytes; genome reduction; leucoplast; non-photosynthesis; plastid genome.
Copyright © 2020 Kim, Jeong, Archibald and Shin.
Figures



References
-
- Andersen R. A., Berges J. A., Harrison P. J., Watanabe M. M. (2005). “Appendix A. Recipes for freshwater and seawater media” in Algal culturing techniques. Ed. Andersen R. A. (Elsevier Academic Press; ), 429–538. 10.1016/B978-012088426-1/50027-5 - DOI
-
- Andersen R. A., Graf L., Malakhov Y., Yoon H. S. (2017). Rediscovery of the Ochromonas type species Ochromonas triangulata (Chrysophyceae) from its type locality (Lake Veysove, Donetsk region, Ukraine). Phycologia 56, 591–604. 10.2216/17-15.1 - DOI
-
- Barrett C. F., Freudenstein J. V., Li J., Mayfield-Jones D. R., Perez L., Pires J. C., et al. (2014). Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Mol. Biol. Evol. 31, 3095–3112. 10.1093/molbev/msu252 - DOI - PubMed
LinkOut - more resources
Full Text Sources

