Partial Amniote Sex Chromosomal Linkage Homologies Shared on Snake W Sex Chromosomes Support the Ancestral Super-Sex Chromosome Evolution in Amniotes
- PMID: 33014016
- PMCID: PMC7461878
- DOI: 10.3389/fgene.2020.00948
Partial Amniote Sex Chromosomal Linkage Homologies Shared on Snake W Sex Chromosomes Support the Ancestral Super-Sex Chromosome Evolution in Amniotes
Abstract
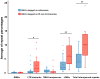
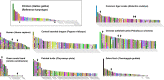
Squamate reptile chromosome 2 (SR2) is thought to be an important remnant of an ancestral amniote super-sex chromosome, but a recent study showed that the Siamese cobra W sex chromosome is also a part of this larger ancestral chromosome. To confirm the existence of an ancestral amniote super-sex chromosome and understand the mechanisms of amniote sex chromosome evolution, chromosome maps of two snake species [Russell's viper: Daboia russelii (DRU) and the common tiger snake: Notechis scutatus (NSC)] were constructed using bacterial artificial chromosomes (BACs) derived from chicken and zebra finch libraries containing amniote sex chromosomal linkages. Sixteen BACs were mapped on the W sex chromosome of DRU and/or NSC, suggesting that these BACs contained a common genomic region shared with the W sex chromosome of these snakes. Two of the sixteen BACs were co-localized to DRU2 and NSC2, corresponding to SR2. Prediction of genomic content from all BACs mapped on snake W sex chromosomes revealed a large proportion of long interspersed nuclear element (LINE) and short interspersed nuclear element (SINE) retrotransposons. These results led us to predict that amplification of LINE and SINE may have occurred on snake W chromosomes during evolution. Genome compartmentalization, such as transposon amplification, might be the key factor influencing chromosome structure and differentiation. Multiple sequence alignments of all BACs mapped on snake W sex chromosomes did not reveal common sequences. Our findings indicate that the SR2 and snake W sex chromosomes may have been part of a larger ancestral amniote super-sex chromosome, and support the view of sex chromosome evolution as a colorful myriad of situations and trajectories in which many diverse processes are in action.
Keywords: BAC; chromosome map; retrotransposon; snake; super-sex chromosome.
Copyright © 2020 Singchat, Ahmad, Sillapaprayoon, Muangmai, Duengkae, Peyachoknagul, O’Connor, Griffin and Srikulnath.
Figures



Similar articles
-
Remnant of Unrelated Amniote Sex Chromosomal Linkage Sharing on the Same Chromosome in House Gecko Lizards, Providing a Better Understanding of the Ancestral Super-Sex Chromosome.Cells. 2021 Nov 1;10(11):2969. doi: 10.3390/cells10112969. Cells. 2021. PMID: 34831192 Free PMC article.
-
Do sex chromosomes of snakes, monitor lizards, and iguanian lizards result from multiple fission of an "ancestral amniote super-sex chromosome"?Chromosome Res. 2020 Jun;28(2):209-228. doi: 10.1007/s10577-020-09631-4. Epub 2020 May 1. Chromosome Res. 2020. PMID: 32358743
-
Chromosome map of the Siamese cobra: did partial synteny of sex chromosomes in the amniote represent "a hypothetical ancestral super-sex chromosome" or random distribution?BMC Genomics. 2018 Dec 17;19(1):939. doi: 10.1186/s12864-018-5293-6. BMC Genomics. 2018. PMID: 30558533 Free PMC article.
-
Snake W Sex Chromosome: The Shadow of Ancestral Amniote Super-Sex Chromosome.Cells. 2020 Oct 31;9(11):2386. doi: 10.3390/cells9112386. Cells. 2020. PMID: 33142713 Free PMC article. Review.
-
Tracing the evolution of amniote chromosomes.Chromosoma. 2014 Jun;123(3):201-16. doi: 10.1007/s00412-014-0456-y. Epub 2014 Mar 25. Chromosoma. 2014. PMID: 24664317 Free PMC article. Review.
Cited by
-
Existence of Bov-B LINE Retrotransposons in Snake Lineages Reveals Recent Multiple Horizontal Gene Transfers with Copy Number Variation.Genes (Basel). 2020 Oct 22;11(11):1241. doi: 10.3390/genes11111241. Genes (Basel). 2020. PMID: 33105659 Free PMC article.
-
Remnant of Unrelated Amniote Sex Chromosomal Linkage Sharing on the Same Chromosome in House Gecko Lizards, Providing a Better Understanding of the Ancestral Super-Sex Chromosome.Cells. 2021 Nov 1;10(11):2969. doi: 10.3390/cells10112969. Cells. 2021. PMID: 34831192 Free PMC article.
-
Conservation of Major Satellite DNAs in Snake Heterochromatin.Animals (Basel). 2023 Jan 17;13(3):334. doi: 10.3390/ani13030334. Animals (Basel). 2023. PMID: 36766223 Free PMC article.
-
MicrosatNavigator: exploring nonrandom distribution and lineage-specificity of microsatellite repeat motifs on vertebrate sex chromosomes across 186 whole genomes.Chromosome Res. 2023 Sep 30;31(4):29. doi: 10.1007/s10577-023-09738-4. Chromosome Res. 2023. PMID: 37775555
-
Why Do Some Vertebrates Have Microchromosomes?Cells. 2021 Aug 24;10(9):2182. doi: 10.3390/cells10092182. Cells. 2021. PMID: 34571831 Free PMC article. Review.
References
-
- Ahmad S. F., Singchat W., Jehangir M., Panthum T., Srikulnath K. (2020). Consequence of paradigm shift with repeat landscapes in reptiles: powerful facilitators of chromosomal rearrangements for diversity and evolution (running title: genomic impact of repeats on chromosomal dynamics in reptiles). Genes 11:827. 10.3390/genes11070827 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

