A Urine Metabonomics Study of Rat Bladder Cancer by Combining Gas Chromatography-Mass Spectrometry with Random Forest Algorithm
- PMID: 33014064
- PMCID: PMC7525317
- DOI: 10.1155/2020/8839215
A Urine Metabonomics Study of Rat Bladder Cancer by Combining Gas Chromatography-Mass Spectrometry with Random Forest Algorithm
Abstract
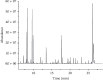
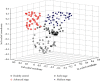
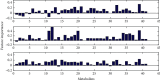
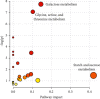
A urine metabolomics study based on gas chromatography-mass spectrometry (GC-MS) and multivariate statistical analysis was applied to distinguish rat bladder cancer. Urine samples with different stages were collected from animal models, i.e., the early stage, medium stage, and advanced stage of the bladder cancer model group and healthy group. After resolving urea with urease, the urine samples were extracted with methanol and, then, derived with N, O-Bis(trimethylsilyl) trifluoroacetamide and trimethylchlorosilane (BSTFA + TMCS, 99 : 1, v/v), before analyzed by GC-MS. Three classification models, i.e., healthy control vs. early- and middle-stage groups, healthy control vs. advanced-stage group, and early- and middle-stage groups vs. advanced-stage group, were established to analyze these experimental data by using Random Forests (RF) algorithm, respectively. The classification results showed that combining random forest algorithm with metabolites characters, the differences caused by the progress of disease could be effectively exhibited. Our results showed that glyceric acid, 2, 3-dihydroxybutanoic acid, N-(oxohexyl)-glycine, and D-turanose had higher contributions in classification of different groups. The pathway analysis results showed that these metabolites had relationships with starch and sucrose, glycine, serine, threonine, and galactose metabolism. Our study results suggested that urine metabolomics was an effective approach for disease diagnosis.
Copyright © 2020 Mengchan Fang et al.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Mahendran N., xie G.-X., Jia W. Metabonomics of human colorectal cancer: new approaches for early diagnosis and biomarker discovery. Journal of Proteome Research. 2014;13(9):3857–3870. - PubMed
-
- Xie W., Zhang W., Ren J., et al. Metabonomics indicates inhibition of fatty acid synthesis, β-oxidation, and tricarboxylic acid cycle in triclocarban-induced cardiac metabolic alterations in male mice. Journal of Agricultural and Food Chemistry. 2018;66(6):1533–1542. doi: 10.1021/acs.jafc.7b05220. - DOI - PubMed
-
- Li C., Sturm S. Analytical aspects of plant metabolite profiling platforms: current standings and future aims. Journal of Proteome Research. 2007;6(2):480–497. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

