Diagnostic and Prognostic Potentials of Long Noncoding RNA ELF3-AS1 in Glioma Patients
- PMID: 33014189
- PMCID: PMC7519982
- DOI: 10.1155/2020/8871746
Diagnostic and Prognostic Potentials of Long Noncoding RNA ELF3-AS1 in Glioma Patients
Abstract
Objective: Accumulating evidence implies that long noncoding RNAs (lncRNAs) play a crucial role in predicting survival for glioma patients. However, the potential function of lncRNA ELF3-antisense RNA 1 (ELF3-AS1) in tumors remained largely unclear. The aim of this study was to explore the expression of lncRNA ELF3-antisense RNA 1 (ELF3-AS1) and evaluate its functions in glioma patients. Patients and Methods. ELF3-AS1 expressions were examined by RT-PCR in 182 pairs of glioma specimens and adjacent normal tissues. The receiver operating characteristic (ROC) curve was performed to estimate the diagnostic value of ELF3-AS1. The chi-square tests were used to examine the associations between ELF3-AS1 expression and the clinicopathological characters. The overall survival (OS) and disease-free survival (DFS) were analyzed by log-rank test, and survival curves were plotted according to Kaplan-Meier. The prognostic value of the ELF3-AS1 expression in glioma patients was further analyzed using univariate and multivariate Cox regression analyses. Loss-of-function assays were performed to determine the potential function of ELF3-AS1 on the proliferation and invasion of glioma cells.
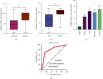
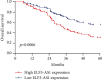
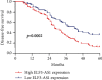
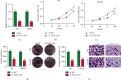
Results: The ELF3-AS1 expression level was significantly higher in glioma specimens compared with adjacent nontumor specimens (p < 0.01). A high expression of ELF3-AS1 was shown to be associated with the WHO grade (p = 0.023) and KPS score (p = 0.012). ROC assays revealed that high ELF3-AS1 expression had an AUC value of 0.8073 (95% CI: 0.7610 to 0.8535) for glioma. Using the Kaplan-Meier analysis, we found that patients with a high ELF3-AS1 expression had significantly poor OS (p = 0.006) and DFS (p = 0.0002). In a multivariate Cox model, we confirmed that ELF3-AS1 expression was an independent poor prognostic factor for glioma patients. The functional assay revealed that knockdown of ELF3-AS1 suppressed the proliferation and invasion of glioma cells.
Conclusions: Our findings confirmed that ELF3-AS1 functions as an oncogene in glioma and indicated that ELF3-AS1 is not only an important prognostic marker but also a potential therapy target for glioma.
Copyright © 2020 Jun-chi Mei et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures
Similar articles
-
LncRNA ELF3-AS1 is a Prognostic Biomarker and Correlated with Immune Infiltrates in Hepatocellular Carcinoma.Can J Gastroenterol Hepatol. 2021 Jul 9;2021:8323487. doi: 10.1155/2021/8323487. eCollection 2021. Can J Gastroenterol Hepatol. 2021. PMID: 34336727 Free PMC article.
-
Analysis of Long Noncoding RNA ZNF667-AS1 as a Potential Biomarker for Diagnosis and Prognosis of Glioma Patients.Dis Markers. 2020 Nov 16;2020:8895968. doi: 10.1155/2020/8895968. eCollection 2020. Dis Markers. 2020. PMID: 33282010 Free PMC article.
-
ELF3-AS1 contributes to gastric cancer progression by binding to hnRNPK and induces thrombocytosis in peripheral blood.Cancer Sci. 2021 Nov;112(11):4553-4569. doi: 10.1111/cas.15104. Epub 2021 Sep 9. Cancer Sci. 2021. PMID: 34418240 Free PMC article.
-
Long Non-Coding RNA AGAP2-AS1: A Comprehensive Overview on Its Biological Functions and Clinical Significances in Human Cancers.Molecules. 2024 Jul 24;29(15):3461. doi: 10.3390/molecules29153461. Molecules. 2024. PMID: 39124865 Free PMC article. Review.
-
Long Noncoding RNA LIFR-AS1: A New Player in Human Cancers.Biomed Res Int. 2022 Jan 13;2022:1590815. doi: 10.1155/2022/1590815. eCollection 2022. Biomed Res Int. 2022. PMID: 35071590 Free PMC article. Review.
Cited by
-
Evolving Landscape of Long Non-coding RNAs in Cerebrospinal Fluid: A Key Role From Diagnosis to Therapy in Brain Tumors.Front Cell Dev Biol. 2021 Oct 7;9:737670. doi: 10.3389/fcell.2021.737670. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34692695 Free PMC article. Review.
-
Subtype cluster analysis unveiled the correlation between m6A- and cuproptosis-related lncRNAs and the prognosis, immune microenvironment, and treatment sensitivity of esophageal cancer.Front Immunol. 2025 Feb 17;16:1539630. doi: 10.3389/fimmu.2025.1539630. eCollection 2025. Front Immunol. 2025. PMID: 40034693 Free PMC article.
-
LncRNA ELF3-AS1 is a Prognostic Biomarker and Correlated with Immune Infiltrates in Hepatocellular Carcinoma.Can J Gastroenterol Hepatol. 2021 Jul 9;2021:8323487. doi: 10.1155/2021/8323487. eCollection 2021. Can J Gastroenterol Hepatol. 2021. PMID: 34336727 Free PMC article.
-
Glioblastoma at the crossroads: current understanding and future therapeutic horizons.Signal Transduct Target Ther. 2025 Jul 9;10(1):213. doi: 10.1038/s41392-025-02299-4. Signal Transduct Target Ther. 2025. PMID: 40628732 Free PMC article. Review.
-
The Involvement of Long Non-Coding RNAs in Glioma: From Early Detection to Immunotherapy.Front Immunol. 2022 May 10;13:897754. doi: 10.3389/fimmu.2022.897754. eCollection 2022. Front Immunol. 2022. PMID: 35619711 Free PMC article. Review.
References
-
- Delgado-López P. D., Corrales-García E. M., Martino J., Lastra-Aras E., Dueñas-Polo M. T. Diffuse low-grade glioma: a review on the new molecular classification, natural history and current management strategies. Clinical and Translational Oncology. 2017;19(8):931–944. doi: 10.1007/s12094-017-1631-4. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

