Genome-wide microsatellites and species specific markers in genus Phytophthora revealed through whole genome analysis
- PMID: 33014685
- PMCID: PMC7501370
- DOI: 10.1007/s13205-020-02430-y
Genome-wide microsatellites and species specific markers in genus Phytophthora revealed through whole genome analysis
Abstract
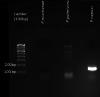
Genome wide microsatellite maps shall support Phytophthora systematics through the development of reliable markers, enabling species discrimination and variability analyses. Whole genome sequences of 17 Phytophthora accessions belonging to 14 species were retrieved from GenBank and the genome-wide microsatellites in each species were mined. A total of 51,200 microsatellites, including dinucleotide to decanucleotide motifs, have been identified across all the species and each one was characterized for uniqueness and repeat number. The P. infestans T30-4 genome had the highest (6873) and P. multivora 3378 had the lowest number of microsatellites (1802). Dinucleotide motifs (63.6%) followed by trinucleotide motifs (30.1%) were most abundant in all the genome. From 14 species, 250 microsatellites which are unique for the respective genomes are detailed along with their primer combinations and product sizes. P. sojae had the highest number of unique microsatellite motifs. Genome wide microsatellite maps for all the 14 Phytophthora species including the chromosome, position, motif, repeat number, forward and reverse primer sequences and expected PCR product size, for every microsatellite are presented. Markers based on the unique microsatellites could be used to identify each species, whereas the ones common to all species could be used to identify the genetic variability. Furthermore, to confirm the results, pure cultures of P. capsici, P. nicotianae and P. palmivora were procured from the Phytophthora Repository, DNA was isolated and the unique markers were screened across the species. The characteristic markers developed have confirmed the genome analysis results.
Keywords: Fungal taxonomy; Genome annotation; Microsatellite markers; Next generation sequencing (NGS) data analysis; Simple sequence repeats (SSR); Species identification.
© King Abdulaziz City for Science and Technology 2020.
Conflict of interest statement
Conflict of interestOn behalf of all authors, the corresponding author states that there is no conflict of interest.
Figures

References
-
- Afandi A, Hieno A, Wibowo A, Subandiyah S, Suga H, Tsuchida K, Kageyama K. Genetic diversity of Phytophthora nicotianae reveals pathogen transmission mode in Japan. J Gen Plant Pathol. 2019;85(3):189–200. doi: 10.1007/s10327-018-00836-4. - DOI
-
- Alconero R, Albuquerque F, Almeyda N, Santiago AG. Phytophthora foot rot of black pepper in Brazil and Puerto Rico. Phytopathology. 1972;62(1):144–148. doi: 10.1094/Phyto-62-144. - DOI
-
- Al-Kaabi S, Mayton H, Al-Jabri M, Al-Maqbali R, Al-Shibli W, Myers K, Fry W, Al-Adawi A. Appearance and diversity of Phytophthora infestans in Oman in 2013–2015. J Plant Pathol. 2020 doi: 10.1007/s42161-020-00587-2. - DOI
-
- Arafa RA, Soliman NEK, Moussa OM, Kamel SM, Shirasawa K. Characterization of Egyptian Phytophthora infestans population using simple sequence repeat markers. J Gen Plant Pathol. 2018;84(2):104–107. doi: 10.1007/s10327-018-0763-x. - DOI
-
- Arafa RA, Kamel SM, Rakha MT, Soliman NEK, Moussa OM, Shirasawa K. Analysis of the lineage of Phytophthora infestans isolates using mating type assay, traditional markers, and next generation sequencing technologies. PLoS ONE. 2020;15(1):e0221604. doi: 10.1371/journal.pone.0221604. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

