Evaluation of Two Methods for the Detection of Third Generation Cephalosporins Resistant Enterobacterales Directly From Positive Blood Cultures
- PMID: 33014900
- PMCID: PMC7516202
- DOI: 10.3389/fcimb.2020.00491
Evaluation of Two Methods for the Detection of Third Generation Cephalosporins Resistant Enterobacterales Directly From Positive Blood Cultures
Abstract
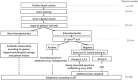
Due to the importance of a rapid determination of patients infected by multidrug resistant bacteria, we evaluated two rapid diagnostic tests for the detection of third-generation cephalosporins (3GC)-resistant Enterobacterales directly from positive blood cultures within 1 h: BL-REDTM (electrochemical method) and β-LACTATM test (chromogenic method). A panel of 150 clinical strains characterized for their resistance profiles (e.g., penicillinases, extended-spectrum beta-lactamases (ESBLs), overproduction of cephalosporinase, carbapenemases, impermeability) was tested. Approximately 100 CFU of each isolate was spiked into sterile blood culture bottles and incubated in a BD BACTECTM FX automated system (Becton Dickinson, USA). Positive blood cultures were examined to parallel testing using the BL-REDTM and β-LACTATM tests and conventional susceptibility method (disc diffusion following EUCAST recommendations). For all phenotypes combined, the sensitivity, specificity, positive predictive value, and negative predictive value in the detection of 3GC resistance were, respectively (i) with BL-REDTM: 45.7, 100, 100, and 54.2% and (ii) with β-LACTATM test: 52.2, 100, 100, and 56.9%. The positivity of tests allows to adapt antibiotic treatment whereas the negative result requires other tests. Moreover, these tests detect most Ambler class A-producing Enterobacterales (KPC, ESBL, extended-spectrum OXY) with sensitivities and specificities of 87.5 and 99% for BL-REDTM, respectively and both 100% for β-LACTATM test (47/47 isolates). These two rapid tests failed to detect AmpC overexpressed (sensitivities of 2.7% for BL-REDTM and 0% for β-LACTATM test) and Ambler class B-producing Enterobacterales (sensitivities of 40% for both tests) notably strains without ESBLs associated (sensitivities of 0% for both tests). BL-REDTM and β-LACTATM tests are easy-to-use and mainly attractive when a positive result is obtained notably to detect most of the Ambler class A-producing Enterobacterales in <1 h after the positivity of the blood culture, allowing a rapid adaptation of the antibiotic therapy in patients.
Keywords: BL-REDTM test; bacteremia; blood cultures; extended-spectrum beta-lactamases; multidrug resistance; third-generation cephalosporins; β-LACTATM test.
Copyright © 2020 Durand, Boudet, Lavigne and Pantel.
Figures
Similar articles
-
Evaluation of Rapid Immunochromatographic Tests for the Direct Detection of Extended Spectrum Beta-Lactamases and Carbapenemases in Enterobacterales Isolated from Positive Blood Cultures.Microbiol Spectr. 2021 Dec 22;9(3):e0078521. doi: 10.1128/Spectrum.00785-21. Epub 2021 Dec 8. Microbiol Spectr. 2021. PMID: 34878297 Free PMC article.
-
Performance of the Accelerate Pheno™ system for identification and antimicrobial susceptibility testing of a panel of multidrug-resistant Gram-negative bacilli directly from positive blood cultures.J Antimicrob Chemother. 2018 Jun 1;73(6):1546-1552. doi: 10.1093/jac/dky032. J Antimicrob Chemother. 2018. PMID: 29474660
-
A rapid and inexpensive protocol to screen for third generation cephalosporin-resistant and non-fermenting Gram-negative rods directly in positive blood cultures.Diagn Microbiol Infect Dis. 2021 Oct;101(2):115428. doi: 10.1016/j.diagmicrobio.2021.115428. Epub 2021 May 16. Diagn Microbiol Infect Dis. 2021. PMID: 34174522
-
Cefiderocol: A Siderophore Cephalosporin with Activity Against Carbapenem-Resistant and Multidrug-Resistant Gram-Negative Bacilli.Drugs. 2019 Feb;79(3):271-289. doi: 10.1007/s40265-019-1055-2. Drugs. 2019. PMID: 30712199 Review.
-
Phenotypic detection of extended-spectrum beta-lactamase production in Enterobacteriaceae: review and bench guide.Clin Microbiol Infect. 2008 Jan;14 Suppl 1:90-103. doi: 10.1111/j.1469-0691.2007.01846.x. Clin Microbiol Infect. 2008. PMID: 18154532 Review.
Cited by
-
Detection of Multidrug-Resistant Enterobacterales-From ESBLs to Carbapenemases.Antibiotics (Basel). 2021 Sep 21;10(9):1140. doi: 10.3390/antibiotics10091140. Antibiotics (Basel). 2021. PMID: 34572722 Free PMC article. Review.
-
Antimicrobial Susceptibility of Community-Acquired Urine Bacterial Isolates in French Amazonia.Am J Trop Med Hyg. 2023 Apr 3;108(5):927-935. doi: 10.4269/ajtmh.22-0242. Print 2023 May 3. Am J Trop Med Hyg. 2023. PMID: 37011893 Free PMC article.
References
-
- Buchan B. W., Ginocchio C. C., Manii R., Cavagnolo R., Pancholi P., Swyers L., et al. . (2013). Multiplex identification of Gram-positive bacteria and resistance determinants directly from positive blood culture broths: evaluation of an automated microarray-based nucleic acid test. PLoS Med. 10:e1001478. 10.1371/journal.pmed.1001478 - DOI - PMC - PubMed
-
- Compain F., Bensekhri H., Rostane H., Mainardi J. L., Lavollay M. (2015). β LACTA test for rapid detection of Enterobacteriaceae resistant to third-generation cephalosporins from positive blood cultures using briefly incubated solid medium cultures. J. Med. Microbiol. 64, 1256–1259. 10.1099/jmm.0.000157 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources