Bacterial Genome Wide Association Studies (bGWAS) and Transcriptomics Identifies Cryptic Antimicrobial Resistance Mechanisms in Acinetobacter baumannii
- PMID: 33014966
- PMCID: PMC7493718
- DOI: 10.3389/fpubh.2020.00451
Bacterial Genome Wide Association Studies (bGWAS) and Transcriptomics Identifies Cryptic Antimicrobial Resistance Mechanisms in Acinetobacter baumannii
Abstract
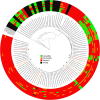
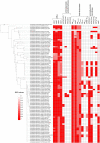
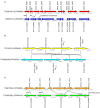
Antimicrobial resistance (AMR) in the nosocomial pathogen, Acinetobacter baumannii, is becoming a serious public health threat. While some mechanisms of AMR have been reported, understanding novel mechanisms of resistance is critical for identifying emerging resistance. One of the first steps in identifying novel AMR mechanisms is performing genotype/phenotype association studies; however, performing these studies is complicated by the plastic nature of the A. baumannii pan-genome. In this study, we compared the antibiograms of 12 antimicrobials associated with multiple drug families for 84 A. baumannii isolates, many isolated in Arizona, USA. in silico screening of these genomes for known AMR mechanisms failed to identify clear correlations for most drugs. We then performed a bacterial genome wide association study (bGWAS) looking for associations between all possible 21-mers; this approach generally failed to identify mechanisms that explained the resistance phenotype. In order to decrease the genomic noise associated with population stratification, we compared four phylogenetically-related pairs of isolates with differing susceptibility profiles. RNA-Sequencing (RNA-Seq) was performed on paired isolates and differentially-expressed genes were identified. In these isolate pairs, five different potential mechanisms were identified, highlighting the difficulty of broad AMR surveillance in this species. To verify and validate differential expression, amplicon sequencing was performed. These results suggest that a diagnostic platform based on gene expression rather than genomics alone may be beneficial in certain surveillance efforts. The implementation of such advanced diagnostics coupled with increased AMR surveillance will potentially improve A. baumannii infection treatment and patient outcomes.
Keywords: AMR; acinetobacter; bioinformatics; genomics; transcriptomics.
Copyright © 2020 Roe, Williamson, Vazquez, Kyger, Valentine, Bowers, Phillips, Harrison, Driebe, Engelthaler and Sahl.
Figures
Similar articles
-
Large-scale genomic analysis reveals significant role of insertion sequences in antimicrobial resistance of Acinetobacter baumannii.mBio. 2025 Mar 12;16(3):e0285224. doi: 10.1128/mbio.02852-24. Epub 2025 Feb 20. mBio. 2025. PMID: 39976435 Free PMC article.
-
Phenotypic and WGS-derived antimicrobial resistance profiles of clinical and non-clinical Acinetobacter baumannii isolates from Germany and Vietnam.Int J Antimicrob Agents. 2020 Oct;56(4):106127. doi: 10.1016/j.ijantimicag.2020.106127. Epub 2020 Aug 1. Int J Antimicrob Agents. 2020. PMID: 32750418
-
WGS based analysis of acquired antimicrobial resistance in human and non-human Acinetobacter baumannii isolates from a German perspective.BMC Microbiol. 2021 Jul 10;21(1):210. doi: 10.1186/s12866-021-02270-7. BMC Microbiol. 2021. PMID: 34243717 Free PMC article.
-
Prevalence of Acinetobacter baumannii in Saudi Arabia: risk factors, antimicrobial resistance patterns and mechanisms of carbapenem resistance.Ann Clin Microbiol Antimicrob. 2019 Jan 3;18(1):1. doi: 10.1186/s12941-018-0301-x. Ann Clin Microbiol Antimicrob. 2019. PMID: 30606201 Free PMC article. Review.
-
Genomics for antimicrobial resistance-progress and future directions.Antimicrob Agents Chemother. 2025 May 7;69(5):e0108224. doi: 10.1128/aac.01082-24. Epub 2025 Apr 14. Antimicrob Agents Chemother. 2025. PMID: 40227048 Free PMC article. Review.
Cited by
-
Making sense of drug-efflux transporters in the physiological environment.Curr Opin Microbiol. 2022 Oct;69:102179. doi: 10.1016/j.mib.2022.102179. Epub 2022 Jul 23. Curr Opin Microbiol. 2022. PMID: 35882103 Free PMC article. Review.
-
Bacterial genome-wide association studies: exploring the genetic variation underlying bacterial phenotypes.Appl Environ Microbiol. 2025 Jun 18;91(6):e0251224. doi: 10.1128/aem.02512-24. Epub 2025 May 16. Appl Environ Microbiol. 2025. PMID: 40377303 Free PMC article. Review.
-
Aminoglycoside resistance genes in early members of the Acinetobacter baumannii ST78A (SMAL, Italian clone) reside in an IS26-bounded island in the chromosome.J Antimicrob Chemother. 2024 May 2;79(5):1014-1018. doi: 10.1093/jac/dkae064. J Antimicrob Chemother. 2024. PMID: 38530861 Free PMC article.
-
Phenotype-Based Threat Assessment.Proc Natl Acad Sci U S A. 2022 Apr 5;119(14):e2112886119. doi: 10.1073/pnas.2112886119. Epub 2022 Apr 1. Proc Natl Acad Sci U S A. 2022. PMID: 35363569 Free PMC article.
-
Update on Multidrug Resistance Efflux Pumps in Acinetobacter spp.Antimicrob Agents Chemother. 2021 Jun 17;65(7):e0051421. doi: 10.1128/AAC.00514-21. Epub 2021 Jun 17. Antimicrob Agents Chemother. 2021. PMID: 33903107 Free PMC article.
References
-
- O'Neill J. Antimicrobial resistance: tackling a crisis for the health and wealth of nations. Rev Antimicrob Resist. (2014) 20:1–16.
-
- WHO Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics (2018).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

