Treatment with Commonly Used Antiretroviral Drugs Induces a Type I/III Interferon Signature in the Gut in the Absence of HIV Infection
- PMID: 33015651
- PMCID: PMC7511692
- DOI: 10.1016/j.xcrm.2020.100096
Treatment with Commonly Used Antiretroviral Drugs Induces a Type I/III Interferon Signature in the Gut in the Absence of HIV Infection
Abstract
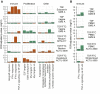
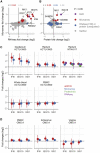
Tenofovir disoproxil fumarate (TDF) and emtricitabine (FTC) are used for HIV treatment and prevention. Previously, we found that topical rectal tenofovir gel caused immunological changes in the mucosa. Here, we assess the effect of oral TDF/FTC in three HIV pre-exposure prophylaxis trials, two with gastrointestinal and one with cervicovaginal biopsies. TDF/FTC induces type I/III interferon-related (IFN I/III) genes in the gastrointestinal tract, but not blood, with strong correlations between the two independent rectal biopsy groups (Spearman r = 0.91) and between the rectum and duodenum (r = 0.81). Gene set testing also indicates stimulation of the type I/III pathways in the ectocervix and of cellular proliferation in the duodenum. mRNA sequencing, digital droplet PCR, proteomics, and immunofluorescence confirm IFN I/III pathway stimulation in the gastrointestinal tract. Thus, oral TDF/FTC stimulates an IFN I/III signature throughout the gut, which could increase antiviral efficacy but also cause chronic immune activation in HIV prevention and treatment settings.
Keywords: ART; HIV; HIV cure; ISG15; antiretroviral treatment; chronic immune activation; gut; interferon; tenofovir.
© 2020 The Author(s).
Conflict of interest statement
J.M.B. is on the advisory boards of Gilead Sciences, Merck, and Janssen. I.M. is the Chief Medical Officer of Orion Biotechnology. All of the other authors declare no competing interests.
Figures



References
-
- Lerner A.M., Eisinger R.W., Fauci A.S. Comorbidities in Persons With HIV: The Lingering Challenge. JAMA. 2019;323:19–20. - PubMed
-
- Alzahrani J., Hussain T., Simar D., Palchaudhuri R., Abdel-Mohsen M., Crowe S.M., Mbogo G.W., Palmer C.S. Inflammatory and immunometabolic consequences of gut dysfunction in HIV: Parallels with IBD and implications for reservoir persistence and non-AIDS comorbidities. EBioMedicine. 2019;46:522–531. - PMC - PubMed
-
- Owens D.K., Davidson K.W., Krist A.H., Barry M.J., Cabana M., Caughey A.B., Curry S.J., Doubeni C.A., Epling J.W., Jr., Kubik M., US Preventive Services Task Force Preexposure Prophylaxis for the Prevention of HIV Infection: US Preventive Services Task Force Recommendation Statement. JAMA. 2019;321:2203–2213. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

