A pipeline for precise and efficient genome editing by sgRNA-Cas9 RNPs in Drosophila
- PMID: 33016195
- PMCID: PMC7746241
- DOI: 10.1080/19336934.2020.1832416
A pipeline for precise and efficient genome editing by sgRNA-Cas9 RNPs in Drosophila
Abstract
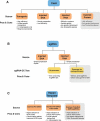
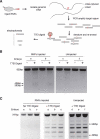
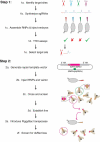
Genome editing via homology-directed repair (HDR) has made possible precise and deliberate modifications to gene sequences. CRISPR/Cas9-mediated HDR is the simplest means to carry this out. However, technical challenges remain to improve efficiency and broaden applicability to any genetic background of Drosophila melanogaster as well as to other Drosophila species. To address these issues, we developed a two-stage marker-assisted strategy in which embryos are injected with RNPs and pre-screened using T7EI. Using sgRNA in complex with recombinant Cas9 protein, we assayed each sgRNA for genome-cutting efficiency. We then conducted HDR using sgRNAs that efficiently cut target genes and the application of a transformation marker that generates RNAi against eyes absent. This allows for screening based on eye morphology rather than colour. These new tools can be used to make a single change or a series of allelic substitutions in a region of interest, or to create additional genetic tools such as balancer chromosomes.
Keywords: CRISPR; Drosophila; gene editing; homology dependent repair.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
-
- Klompe SE, Vo PLH, Halpin-Healy TS, et al. Transposon-encoded CRISPR-Cas systems direct RNA-guided DNA integration. Nature. 2019;571:219–225. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
