Essential role of autophagy in restricting poliovirus infection revealed by identification of an ATG7 defect in a poliomyelitis patient
- PMID: 33016799
- PMCID: PMC8496727
- DOI: 10.1080/15548627.2020.1831800
Essential role of autophagy in restricting poliovirus infection revealed by identification of an ATG7 defect in a poliomyelitis patient
Abstract
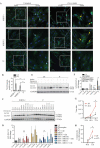
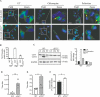
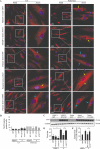
Paralytic poliomyelitis is a rare disease manifestation following poliovirus (PV) infection. The disease determinants remain largely unknown. We used whole exome sequencing to uncover possible contributions of host genetics to the development of disease outcome in humans with poliomyelitis. We identified a patient with a variant in ATG7, an important regulatory gene in the macroautophagy/autophagy pathway. PV infection did not induce a prominent type I interferon response, but rather activated autophagy in neuronal-like cells, and this was essential for viral control. Importantly, virus-induced autophagy was impaired in patient fibroblasts and associated with increased viral burden and enhanced cell death following infection. Lack of ATG7 prevented control of infection in neuronal-like cells, and reconstitution of patient cells with wild-type ATG7 reestablished autophagy-mediated control of infection. Collectively, these data suggest that ATG7 defect contributes to host susceptibility to PV infection and propose autophagy as an unappreciated antiviral effector in viral infection in humans.
Keywords: ATG7; autophagy; host genetics; innate immunity; neuronal-like cells; poliomyelitis; poliovirus; whole exome sequencing.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Nathanson N. The pathogenesis of poliomyelitis: what we don’t know. Adv Virus Res. 2008;71:1–50. - PubMed
-
- Kindberg E, Ax C, Fiore L, et al. Ala67Thr mutation in the poliovirus receptor CD155 is a potential risk factor for vaccine and wild-type paralytic poliomyelitis. J Med Virol. 2009;81(5):933–936. - PubMed
-
- Sabin AB. Pathogenesis of poliomyelitis; reappraisal in the light of new data. Science. 1956;123(3209):1151–1157. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
