mRNA vaccine-induced neoantigen-specific T cell immunity in patients with gastrointestinal cancer
- PMID: 33016924
- PMCID: PMC7598064
- DOI: 10.1172/JCI134915
mRNA vaccine-induced neoantigen-specific T cell immunity in patients with gastrointestinal cancer
Abstract
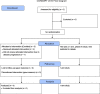
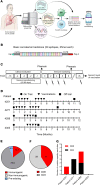
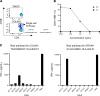
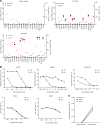
BACKGROUNDTherapeutic vaccinations against cancer have mainly targeted differentiation antigens, cancer-testis antigens, and overexpressed antigens and have thus far resulted in little clinical benefit. Studies conducted by multiple groups have demonstrated that T cells recognizing neoantigens are present in most cancers and offer a specific and highly immunogenic target for personalized vaccination.METHODSWe recently developed a process using tumor-infiltrating lymphocytes to identify the specific immunogenic mutations expressed in patients' tumors. Here, validated, defined neoantigens, predicted neoepitopes, and mutations of driver genes were concatenated into a single mRNA construct to vaccinate patients with metastatic gastrointestinal cancer.RESULTSThe vaccine was safe and elicited mutation-specific T cell responses against predicted neoepitopes not detected before vaccination. Furthermore, we were able to isolate and verify T cell receptors targeting KRASG12D mutation. We observed no objective clinical responses in the 4 patients treated in this trial.CONCLUSIONThis vaccine was safe, and potential future combination of such vaccines with checkpoint inhibitors or adoptive T cell therapy should be evaluated for possible clinical benefit in patients with common epithelial cancers.TRIAL REGISTRATIONPhase I/II protocol (NCT03480152) was approved by the IRB committee of the NIH and the FDA.FUNDINGCenter for Clinical Research, NCI, NIH.
Keywords: Cancer immunotherapy; Oncology; Vaccines.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

