Adaptation of Proteasomes and Lysosomes to Cellular Environments
- PMID: 33019542
- PMCID: PMC7600607
- DOI: 10.3390/cells9102221
Adaptation of Proteasomes and Lysosomes to Cellular Environments
Abstract
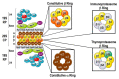
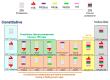
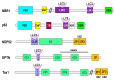
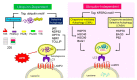
Protein degradation is important for proper cellular physiology as it removes malfunctioning proteins or can provide a source for energy. Proteasomes and lysosomes, through the regulatory particles or adaptor proteins, respectively, recognize proteins destined for degradation. These systems have developed mechanisms to allow adaptation to the everchanging environment of the cell. While the complex recognition of proteins to be degraded is somewhat understood, the mechanisms that help switch the proteasomal regulatory particles or lysosomal adaptor proteins to adjust to the changing landscape of degrons, during infections or inflammation, still need extensive exploration. Therefore, this review is focused on describing the protein degradation systems and the possible sensors that may trigger the rapid adaptation of the protein degradation machinery.
Keywords: aggresome; autophagy; core particle; endosome; protein degradation; regulatory particle.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

