An All-In-One Transcriptome-Based Assay to Identify Therapy-Guiding Genomic Aberrations in Nonsmall Cell Lung Cancer Patients
- PMID: 33019710
- PMCID: PMC7650834
- DOI: 10.3390/cancers12102843
An All-In-One Transcriptome-Based Assay to Identify Therapy-Guiding Genomic Aberrations in Nonsmall Cell Lung Cancer Patients
Abstract
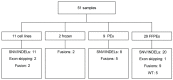
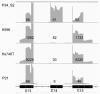
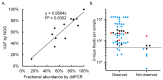
The number of genomic aberrations known to be relevant in making therapeutic decisions for non-small cell lung cancer patients has increased in the past decade. Multiple molecular tests are required to reliably establish the presence of these aberrations, which is challenging because available tissue specimens are generally small. To optimize diagnostic testing, we developed a transcriptome-based next-generation sequencing (NGS) assay based on single primed enrichment technology. We interrogated 11 cell lines, two patient-derived frozen biopsies, nine pleural effusion, and 29 formalin-fixed paraffin-embedded (FFPE) samples. All clinical samples were selected based on previously identified mutations at the DNA level in EGFR, KRAS, ALK, PIK3CA, BRAF, AKT1, MET, NRAS, or ROS1 at the DNA level, or fusion genes at the chromosome level, or by aberrant protein expression of ALK, ROS1, RET, and NTRK1. A successful analysis is dependent on the number of unique reads and the RNA quality, as indicated by the DV200 value. In 27 out of 51 samples with >50 K unique reads and a DV200 >30, all 19 single nucleotide variants (SNVs)/small insertions and deletions (INDELs), three MET exon 14 skipping events, and 13 fusion gene transcripts were detected at the RNA level, giving a test accuracy of 100%. In summary, this lung-cancer-specific all-in-one transcriptome-based assay for the simultaneous detection of mutations and fusion genes is highly sensitive.
Keywords: RNA sequencing; exon skipping; gene fusion; mutation; non-small cell lung cancer.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
Similar articles
-
Validation of a DNA-Based Next-Generation Sequencing Test for Molecular Diagnostic Variant and Fusion Detection in Formalin-Fixed, Paraffin-Embedded Tissue Specimens and Liquid Biopsy Plasma/Cell-Free DNA Samples.J Mol Diagn. 2022 Jul;24(7):784-802. doi: 10.1016/j.jmoldx.2022.04.001. J Mol Diagn. 2022. PMID: 35787794
-
Molecular profiling of small cell lung cancer in a Japanese cohort.Lung Cancer. 2014 May;84(2):139-44. doi: 10.1016/j.lungcan.2014.02.013. Epub 2014 Mar 3. Lung Cancer. 2014. PMID: 24657128
-
Multiplex Diagnosis of Oncogenic Fusion and MET Exon Skipping by Molecular Counting Using Formalin-Fixed Paraffin Embedded Lung Adenocarcinoma Tissues.J Thorac Oncol. 2016 Feb;11(2):203-12. doi: 10.1016/j.jtho.2015.10.005. Epub 2015 Dec 19. J Thorac Oncol. 2016. PMID: 26845116
-
Management of advanced non-small cell lung cancers with known mutations or rearrangements: latest evidence and treatment approaches.Ther Adv Respir Dis. 2016 Apr;10(2):113-29. doi: 10.1177/1753465815617871. Epub 2015 Nov 30. Ther Adv Respir Dis. 2016. PMID: 26620497 Free PMC article. Review.
-
A 2015 update on predictive molecular pathology and its role in targeted cancer therapy: a review focussing on clinical relevance.Cancer Gene Ther. 2015 Sep;22(9):417-30. doi: 10.1038/cgt.2015.39. Epub 2015 Sep 11. Cancer Gene Ther. 2015. PMID: 26358176 Review.
Cited by
-
Analytic and Clinical Validation of a Pan-Cancer NGS Liquid Biopsy Test for the Detection of Copy Number Amplifications, Fusions and Exon Skipping Variants.Diagnostics (Basel). 2022 Mar 17;12(3):729. doi: 10.3390/diagnostics12030729. Diagnostics (Basel). 2022. PMID: 35328282 Free PMC article.
-
New Therapeutic Strategies for Lung Cancer.Cancers (Basel). 2021 Apr 16;13(8):1937. doi: 10.3390/cancers13081937. Cancers (Basel). 2021. PMID: 33923765 Free PMC article.
-
MET alterations detection platforms and clinical implications in solid tumors: a comprehensive review of literature.Ther Adv Med Oncol. 2024 Jan 18;16:17588359231221910. doi: 10.1177/17588359231221910. eCollection 2024. Ther Adv Med Oncol. 2024. PMID: 38249331 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

