Genetic Rescue of X-Linked Retinoschisis Mouse (Rs1-/y) Retina Induces Quiescence of the Retinal Microglial Inflammatory State Following AAV8- RS1 Gene Transfer and Identifies Gene Networks Underlying Retinal Recovery
- PMID: 33019822
- PMCID: PMC8312029
- DOI: 10.1089/hum.2020.213
Genetic Rescue of X-Linked Retinoschisis Mouse (Rs1-/y) Retina Induces Quiescence of the Retinal Microglial Inflammatory State Following AAV8- RS1 Gene Transfer and Identifies Gene Networks Underlying Retinal Recovery
Abstract
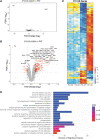
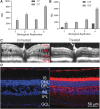
To understand RS1 gene interaction networks in the X-linked retinoschisis (XLRS) mouse retina (Rs1-/y), we analyzed the transcriptome by RNA sequencing before and after in vivo expression of exogenous retinoschisin (RS1) gene delivered by AAV8. RS1 is a secreted cell adhesion protein that is critical for maintaining structural lamination and synaptic integrity of the neural retina. RS1 loss-of-function mutations cause XLRS disease in young boys and men, with splitting ("schisis") of retinal layers and synaptic dysfunction that cause progressive vision loss with age. Analysis of differential gene expression profiles and pathway enrichment analysis of Rs1-KO (Rs1-/y) retina identified cell surface receptor signaling and positive regulation of cell adhesion as potential RS1 gene interaction networks. Most importantly, it also showed massive dysregulation of immune response genes at early age, with characteristics of a microglia-driven proinflammatory state. Delivery of AAV8-RS1 primed the Rs1-KO retina toward structural and functional recovery. The disease transcriptome transitioned toward a recovery phase with upregulation of genes implicated in wound healing, anatomical structure (camera type eye) development, metabolic pathways, and collagen IV networks that provide mechanical stability to basement membrane. AAV8-RS1 expression also attenuated the microglia gene signatures to low levels toward immune quiescence. This study is among the first to identify RS1 gene interaction networks that underlie retinal structural and functional recovery after RS1 gene therapy. Significantly, it also shows that providing wild-type RS1 gene function caused the retina immune status to transition from a degenerative inflammatory phenotype toward immune quiescence, even though the transgene is not directly linked to microglia function. This study indicates that inhibition of microglial proinflammatory responses is an integral part of therapeutic rescue in XLRS gene therapy, and gene therapy might realize its full potential if delivered before microglia activation and photoreceptor cell death. Clinical Trials. gov Identifier NTC 02317887.
Keywords: AAV8-retinoschisin; RNA-seq; X-linked retinoschisis; gene therapy; immune quiescence; microglia activation.
Conflict of interest statement
No competing financial interests exist.
Figures


References
-
- Gehrig AE, Warneke-Wittstock R, Sauer CG, et al. Isolation and characterization of the murine X-linked juvenile retinoschisis (Rs1h) gene. Mamm Genome 1999;10:303–307 - PubMed
-
- Sauer CG, Gehrig A, Warneke-Wittstock R, et al. Positional cloning of the gene associated with X-linked juvenile retinoschisis. Nat Genet 1997;17:164–170 - PubMed
-
- Rao P, Dedania VS, Drenser KA. Congenital X-linked retinoschisis: an updated clinical review. Asia Pac J Ophthalmol (Phila) 2018;7:169–175 - PubMed
-
- Tantri A, Vrabec TR, Cu-Unjieng A, et al. X-linked retinoschisis: a clinical and molecular genetic review. Surv Ophthalmol 2004;49:214–230 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
