Real-time observation of ligand-induced allosteric transitions in a PDZ domain
- PMID: 33020277
- PMCID: PMC7585012
- DOI: 10.1073/pnas.2012999117
Real-time observation of ligand-induced allosteric transitions in a PDZ domain
Abstract
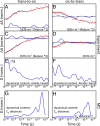
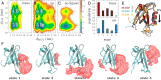
While allostery is of paramount importance for protein regulation, the underlying dynamical process of ligand (un)binding at one site, resulting time evolution of the protein structure, and change of the binding affinity at a remote site are not well understood. Here the ligand-induced conformational transition in a widely studied model system of allostery, the PDZ2 domain, is investigated by transient infrared spectroscopy accompanied by molecular dynamics simulations. To this end, an azobenzene-derived photoswitch is linked to a peptide ligand in a way that its binding affinity to the PDZ2 domain changes upon switching, thus initiating an allosteric transition in the PDZ2 domain protein. The subsequent response of the protein, covering four decades of time, ranging from ∼1 ns to ∼μs, can be rationalized by a remodeling of its rugged free-energy landscape, with very subtle shifts in the populations of a small number of structurally well-defined states. It is proposed that structurally and dynamically driven allostery, often discussed as limiting scenarios of allosteric communication, actually go hand-in-hand, allowing the protein to adapt its free-energy landscape to incoming signals.
Keywords: PDZ domains; allostery; molecular dynamics simulations; transient infrared spectroscopy.
Conflict of interest statement
The authors declare no competing interest.
Figures
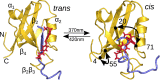


References
-
- Changeux J. P., Allostery and the Monod-Wyman-changeux model after 50 years. Annu. Rev. Biophys. 41, 103–133 (2012). - PubMed
-
- Cooper A., Dryden D. T. F., Allostery without conformational change. Eur. Biophys. J. 11, 103–109 (1984). - PubMed
-
- Fuentes E. J., Gilmore S. A., Mauldin R. V., Lee A. L., Evaluation of energetic and dynamic coupling networks in a PDZ domain protein. J. Mol. Biol. 364, 337–351 (2006). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

