SARS-CoV-2 spike protein predicted to form complexes with host receptor protein orthologues from a broad range of mammals
- PMID: 33020502
- PMCID: PMC7536205
- DOI: 10.1038/s41598-020-71936-5
SARS-CoV-2 spike protein predicted to form complexes with host receptor protein orthologues from a broad range of mammals
Abstract
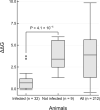
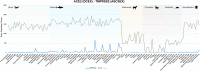
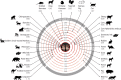
SARS-CoV-2 has a zoonotic origin and was transmitted to humans via an undetermined intermediate host, leading to infections in humans and other mammals. To enter host cells, the viral spike protein (S-protein) binds to its receptor, ACE2, and is then processed by TMPRSS2. Whilst receptor binding contributes to the viral host range, S-protein:ACE2 complexes from other animals have not been investigated widely. To predict infection risks, we modelled S-protein:ACE2 complexes from 215 vertebrate species, calculated changes in the energy of the complex caused by mutations in each species, relative to human ACE2, and correlated these changes with COVID-19 infection data. We also analysed structural interactions to better understand the key residues contributing to affinity. We predict that mutations are more detrimental in ACE2 than TMPRSS2. Finally, we demonstrate phylogenetically that human SARS-CoV-2 strains have been isolated in animals. Our results suggest that SARS-CoV-2 can infect a broad range of mammals, but few fish, birds or reptiles. Susceptible animals could serve as reservoirs of the virus, necessitating careful ongoing animal management and surveillance.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Broad and Differential Animal Angiotensin-Converting Enzyme 2 Receptor Usage by SARS-CoV-2.J Virol. 2020 Aug 31;94(18):e00940-20. doi: 10.1128/JVI.00940-20. Print 2020 Aug 31. J Virol. 2020. PMID: 32661139 Free PMC article.
-
Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2.J Med Virol. 2020 Jun;92(6):595-601. doi: 10.1002/jmv.25726. Epub 2020 Mar 11. J Med Virol. 2020. PMID: 32100877 Free PMC article.
-
Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus.J Virol. 2020 Mar 17;94(7):e00127-20. doi: 10.1128/JVI.00127-20. Print 2020 Mar 17. J Virol. 2020. PMID: 31996437 Free PMC article.
-
Angiotensin-converting enzyme 2: The old door for new severe acute respiratory syndrome coronavirus 2 infection.Rev Med Virol. 2020 Sep;30(5):e2122. doi: 10.1002/rmv.2122. Epub 2020 Jun 30. Rev Med Virol. 2020. PMID: 32602627 Free PMC article. Review.
-
The SARS-CoV-2 Spike Glycoprotein as a Drug and Vaccine Target: Structural Insights into Its Complexes with ACE2 and Antibodies.Cells. 2020 Oct 22;9(11):2343. doi: 10.3390/cells9112343. Cells. 2020. PMID: 33105869 Free PMC article. Review.
Cited by
-
Zoonotic and Reverse Zoonotic Transmissibility of SARS-CoV-2.Virus Res. 2021 Sep;302:198473. doi: 10.1016/j.virusres.2021.198473. Epub 2021 Jun 9. Virus Res. 2021. PMID: 34118360 Free PMC article. Review.
-
Identifying zoonotic origin of SARS-CoV-2 by modeling the binding affinity between Spike receptor-binding domain and host ACE2.bioRxiv [Preprint]. 2020 Sep 11:2020.09.11.293449. doi: 10.1101/2020.09.11.293449. bioRxiv. 2020. Update in: J Proteome Res. 2020 Dec 4;19(12):4844-4856. doi: 10.1021/acs.jproteome.0c00717. PMID: 32935105 Free PMC article. Updated. Preprint.
-
Spread of Mink SARS-CoV-2 Variants in Humans: A Model of Sarbecovirus Interspecies Evolution.Front Microbiol. 2021 Sep 20;12:675528. doi: 10.3389/fmicb.2021.675528. eCollection 2021. Front Microbiol. 2021. PMID: 34616371 Free PMC article. Review.
-
The sound of host-SARS-CoV-2 molecular interactions.Innovation (Camb). 2021 Aug 28;2(3):100126. doi: 10.1016/j.xinn.2021.100126. Epub 2021 Jun 8. Innovation (Camb). 2021. PMID: 34124705 Free PMC article. No abstract available.
-
An update on angiotensin-converting enzyme 2 structure/functions, polymorphism, and duplicitous nature in the pathophysiology of coronavirus disease 2019: Implications for vascular and coagulation disease associated with severe acute respiratory syndrome coronavirus infection.Front Microbiol. 2022 Nov 28;13:1042200. doi: 10.3389/fmicb.2022.1042200. eCollection 2022. Front Microbiol. 2022. PMID: 36519165 Free PMC article. Review.
References
-
- World Health Organisation. COVID-19 situation report—195. Available from: https://www.who.int/docs/default-source/coronaviruse/situation-reports/2....
-
- Shaw LP, Wang AD, Dylus D, et al. The phylogenetic range of bacterial and viral pathogens of vertebrates. Mol Ecol. (2020). 10.1111/mec.15463 - PubMed
MeSH terms
Substances
Grants and funding
- BB/R014892/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/S016007/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/L01257X/1/MRC_/Medical Research Council/United Kingdom
- BB/R01356X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/H018409/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 203780/Z/16/A/WT_/Wellcome Trust/United Kingdom
- BB/S020039/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- MR/P007597/1/MRC_/Medical Research Council/United Kingdom
- BB/T002735/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 104960/Z/14/Z/WT_/Wellcome Trust/United Kingdom
- BB/S020144/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/R009597/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- NH/12/2/29427/BHF_/British Heart Foundation/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

