The immune receptor CD300e negatively regulates T cell activation by impairing the STAT1-dependent antigen presentation
- PMID: 33020563
- PMCID: PMC7536427
- DOI: 10.1038/s41598-020-73552-9
The immune receptor CD300e negatively regulates T cell activation by impairing the STAT1-dependent antigen presentation
Abstract
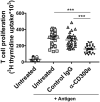
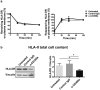
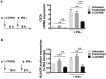
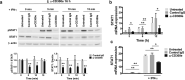
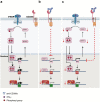
CD300e is a surface receptor, expressed by myeloid cells, involved in the tuning of immune responses. CD300e engagement was reported to provide the cells with survival signals, to trigger the expression of activation markers and the release of pro-inflammatory cytokines. Hence, CD300e is considered an immune activating receptor. In this study, we demonstrate that the ligation of CD300e in monocytes hampers the expression of the human leukocyte antigen (HLA) class II, affecting its synthesis. This effect, which is associated with the transcription impairment of the signal transducer and activator of transcription 1 (STAT1), overcomes the capacity of interferon gamma (IFN-γ) to promote the expression of the antigen-presenting molecules. Importantly, the decreased expression of HLA-II on the surface of CD300e-activated monocytes negatively impacts their capacity to activate T cells in an antigen-specific manner. Notably, unlike in vitro- differentiated macrophages which do not express CD300e, the immune receptor is expressed by tissue macrophages. Taken together, our findings argue against the possibility that this molecule should be considered an activating immune receptor sensu stricto. Moreover, our results support the notion that CD300e might be a new player in the regulation of the expansion of T cell-mediated responses.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

