A major quantitative trait locus on chromosome A9, BnaPh1, controls homoeologous recombination in Brassica napus
- PMID: 33020949
- PMCID: PMC7984352
- DOI: 10.1111/nph.16986
A major quantitative trait locus on chromosome A9, BnaPh1, controls homoeologous recombination in Brassica napus
Abstract
Ensuring faithful homologous recombination in allopolyploids is essential to maintain optimal fertility of the species. Variation in the ability to control aberrant pairing between homoeologous chromosomes in Brassica napus has been identified. The current study exploited the extremes of such variation to identify genetic factors that differentiate newly resynthesised B. napus, which is inherently unstable, and established B. napus, which has adapted to largely control homoeologous recombination. A segregating B. napus mapping population was analysed utilising both cytogenetic observations and high-throughput genotyping to quantify the levels of homoeologous recombination. Three quantitative trait loci (QTL) were identified that contributed to the control of homoeologous recombination in the important oilseed crop B. napus. One major QTL on BnaA9 contributed between 32 and 58% of the observed variation. This study is the first to assess homoeologous recombination and map associated QTLs resulting from deviations in normal pairing in allotetraploid B. napus. The identified QTL regions suggest candidate meiotic genes that could be manipulated in order to control this important trait and further allow the development of molecular markers to utilise this trait to exploit homoeologous recombination in a crop.
Keywords: Brassica; cytogenetics; homoeologous recombination; meiosis; polyploidy.
© 2020 Her Majesty the Queen in Right of Canada. New Phytologist © 2020 New Phytologist Foundation.
Figures


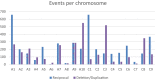
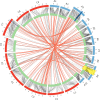
Comment in
-
Homoeologous exchanges in allopolyploids: how Brassica napus established self-control!New Phytol. 2021 Mar;229(6):3041-3043. doi: 10.1111/nph.17222. New Phytol. 2021. PMID: 33616960 No abstract available.
References
-
- Basten CJ, Weir BS, Zeng S‐B. 1999. QTL Cartographer. A reference manual and tutorial for QTL Mapping. Raleigh, NC, USA: Department of Statistics, North Carolina State University.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

