HIV-1 infection activates endogenous retroviral promoters regulating antiviral gene expression
- PMID: 33021676
- PMCID: PMC7641743
- DOI: 10.1093/nar/gkaa832
HIV-1 infection activates endogenous retroviral promoters regulating antiviral gene expression
Abstract
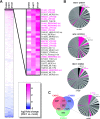
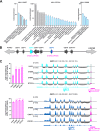
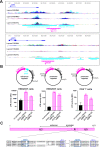
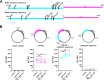
Although endogenous retroviruses (ERVs) are known to harbor cis-regulatory elements, their role in modulating cellular immune responses remains poorly understood. Using an RNA-seq approach, we show that several members of the ERV9 lineage, particularly LTR12C elements, are activated upon HIV-1 infection of primary CD4+ T cells. Intriguingly, HIV-1-induced ERVs harboring transcription start sites are primarily found in the vicinity of immunity genes. For example, HIV-1 infection activates LTR12C elements upstream of the interferon-inducible genes GBP2 and GBP5 that encode for broad-spectrum antiviral factors. Reporter assays demonstrated that these LTR12C elements drive gene expression in primary CD4+ T cells. In line with this, HIV-1 infection triggered the expression of a unique GBP2 transcript variant by activating a cryptic transcription start site within LTR12C. Furthermore, stimulation with HIV-1-induced cytokines increased GBP2 and GBP5 expression in human cells, but not in macaque cells that naturally lack the GBP5 gene and the LTR12C element upstream of GBP2. Finally, our findings suggest that GBP2 and GBP5 have already been active against ancient viral pathogens as they suppress the maturation of the extinct retrovirus HERV-K (HML-2). In summary, our findings uncover how human cells can exploit remnants of once-infectious retroviruses to regulate antiviral gene expression.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Johnson W.E. Origins and evolutionary consequences of ancient endogenous retroviruses. Nat. Rev. Microbiol. 2019; 17:355–370. - PubMed
-
- Mi S., Lee X., Li X., Veldman G.M., Finnerty H., Racie L., LaVallie E., Tang X.Y., Edouard P., Howes S. et al. .. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000; 403:785–789. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

