Benchmarking Orthogroup Inference Accuracy: Revisiting Orthobench
- PMID: 33022036
- PMCID: PMC7738749
- DOI: 10.1093/gbe/evaa211
Benchmarking Orthogroup Inference Accuracy: Revisiting Orthobench
Abstract
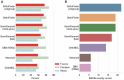
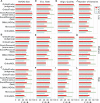
Orthobench is the standard benchmark to assess the accuracy of orthogroup inference methods. It contains 70 expert-curated reference orthogroups (RefOGs) that span the Bilateria and cover a range of different challenges for orthogroup inference. Here, we leveraged improvements in tree inference algorithms and computational resources to reinterrogate these RefOGs and carry out an extensive phylogenetic delineation of their composition. This phylogenetic revision altered the membership of 31 of the 70 RefOGs, with 24 subject to extensive revision and 7 that required minor changes. We further used these revised and updated RefOGs to provide an assessment of the orthogroup inference accuracy of widely used orthogroup inference methods. Finally, we provide an open-source benchmarking suite to support the future development and use of the Orthobench benchmark.
Keywords: benchmark; orthogroup; orthology.
© The Author(s) 2020. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

References
-
- Buchfink B, Xie C, Huson DH.. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 12(1):59–60. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

