Plasma DNA Profile Associated with DNASE1L3 Gene Mutations: Clinical Observations, Relationships to Nuclease Substrate Preference, and In Vivo Correction
- PMID: 33022220
- PMCID: PMC7674998
- DOI: 10.1016/j.ajhg.2020.09.006
Plasma DNA Profile Associated with DNASE1L3 Gene Mutations: Clinical Observations, Relationships to Nuclease Substrate Preference, and In Vivo Correction
Erratum in
-
Plasma DNA Profile Associated with DNASE1L3 Gene Mutations: Clinical Observations, Relationships to Nuclease Substrate Preference, and In Vivo Correction.Am J Hum Genet. 2025 May 1;112(5):1247. doi: 10.1016/j.ajhg.2025.04.001. Am J Hum Genet. 2025. PMID: 40315821 Free PMC article. No abstract available.
Abstract
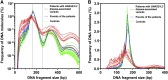
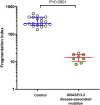
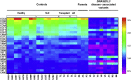
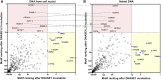
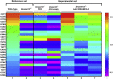
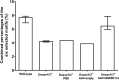
Plasma DNA fragmentomics is an emerging area in cell-free DNA diagnostics and research. In murine models, it has been shown that the extracellular DNase, DNASE1L3, plays a role in the fragmentation of plasma DNA. In humans, DNASE1L3 deficiency causes familial monogenic systemic lupus erythematosus with childhood onset and anti-dsDNA reactivity. In this study, we found that human patients with DNASE1L3 disease-associated gene variations showed aberrations in size and a reduction of a "CC" end motif of plasma DNA. Furthermore, we demonstrated that DNA from DNASE1L3-digested cell nuclei showed a median length of 153 bp with CC motif frequencies resembling plasma DNA from healthy individuals. Adeno-associated virus-based transduction of Dnase1l3 into Dnase1l3-deficient mice restored the end motif profiles to those seen in the plasma DNA of wild-type mice. Our findings demonstrate that DNASE1L3 is an important player in the fragmentation of plasma DNA, which appears to act in a cell-extrinsic manner to regulate plasma DNA size and motif frequency.
Keywords: autoimmune disease; biomarkers; cfDNA; circulating DNA; liquid biopsy; systemic lupus erythematosus.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
R.W.Y.C., M.N., P.J., A.S.H.C., W.P., D.S.C.H., K.C.A.C., R.W.K.C., and Y.M.D.L. are co-inventors on multiple patents in cell-free DNA-based diagnostics. Y.M.D.L. is a scientific co-founder, shareholder, and scientific advisor of Grail. R.W.K.C. and K.C.A.C. are shareholders of Grail. Y.M.D.L., R.W.K.C., and K.C.A.C. are co-founders and shareholders of DRA Limited and the Take2 Group of companies. Y.M.D.L. is an advisor to Decheng Capital. R.W.K.C. is an advisor to Illumina. Y.M.D.L., R.W.K.C., and K.C.A.C. receive royalties from Illumina, Sequenom, DRA, Take2, and Grail.
Figures


References
-
- Chiu R.W.K., Chan K.C.A., Gao Y., Lau V.Y.M., Zheng W., Leung T.Y., Foo C.H.F., Xie B., Tsui N.B.Y., Lun F.M.F. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc. Natl. Acad. Sci. USA. 2008;105:20458–20463. - PMC - PubMed
-
- Chan K.C.A., Woo J.K.S., King A., Zee B.C.Y., Lam W.K.J., Chan S.L., Chu S.W.I., Mak C., Tse I.O.L., Leung S.Y.M. Analysis of plasma Epstein–Barr Virus DNA to screen for nasopharyngeal cancer. N. Engl. J. Med. 2017;377:513–522. - PubMed
-
- Chan K.C.A., Zhang J., Hui A.B.Y., Wong N., Lau T.K., Leung T.N., Lo K.W., Huang D.W.S., Lo Y.M.D. Size distributions of maternal and fetal DNA in maternal plasma. Clin. Chem. 2004;50:88–92. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

