Ubiquitin Phosphorylation at Thr12 Modulates the DNA Damage Response
- PMID: 33022275
- PMCID: PMC7655664
- DOI: 10.1016/j.molcel.2020.09.017
Ubiquitin Phosphorylation at Thr12 Modulates the DNA Damage Response
Abstract
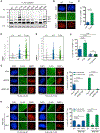
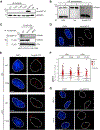
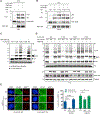
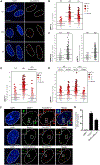
The ubiquitin system regulates the DNA damage response (DDR) by modifying histone H2A at Lys15 (H2AK15ub) and triggering downstream signaling events. Here, we find that phosphorylation of ubiquitin at Thr12 (pUbT12) controls the DDR by inhibiting the function of 53BP1, a key factor for DNA double-strand break repair by non-homologous end joining (NHEJ). Detectable as a chromatin modification on H2AK15ub, pUbT12 accumulates in nuclear foci and is increased upon DNA damage. Mutating Thr12 prevents the removal of ubiquitin from H2AK15ub by USP51 deubiquitinating enzyme, leading to a pronounced accumulation of ubiquitinated chromatin. Chromatin modified by pUbT12 is inaccessible to 53BP1 but permissive to the homologous recombination (HR) proteins RNF169, RAD51, and the BRCA1/BARD1 complex. Phosphorylation of ubiquitin at Thr12 in the chromatin context is a new histone mark, H2AK15pUbT12, that regulates the DDR by hampering the activity of 53BP1 at damaged chromosomes.
Keywords: 53BP1; BRCA1/BARD1; DDR; DNA damage response; DNA repair; H2AK15pUbT12; RAD51; RNF168; RNF169; RNF8; USP51; chromatin ubiquitination; genome stability; histone mark H2AK15ub; pUbT12; phospho-ubiquitin Thr12; ubiquitin phosphorylation.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests H.O. is a shareholder of UbiQ B.V.
Figures

Similar articles
-
BARD1 reads H2A lysine 15 ubiquitination to direct homologous recombination.Nature. 2021 Aug;596(7872):433-437. doi: 10.1038/s41586-021-03776-w. Epub 2021 Jul 28. Nature. 2021. PMID: 34321663
-
Human RNF169 is a negative regulator of the ubiquitin-dependent response to DNA double-strand breaks.J Cell Biol. 2012 Apr 16;197(2):189-99. doi: 10.1083/jcb.201109100. Epub 2012 Apr 9. J Cell Biol. 2012. PMID: 22492721 Free PMC article.
-
53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark.Nature. 2013 Jul 4;499(7456):50-4. doi: 10.1038/nature12318. Epub 2013 Jun 12. Nature. 2013. PMID: 23760478 Free PMC article.
-
Histone Ubiquitination: An Integrative Signaling Platform in Genome Stability.Trends Genet. 2021 Jun;37(6):566-581. doi: 10.1016/j.tig.2020.12.005. Epub 2021 Jan 20. Trends Genet. 2021. PMID: 33485674 Review.
-
An insight into understanding the coupling between homologous recombination mediated DNA repair and chromatin remodeling mechanisms in plant genome: an update.Cell Cycle. 2021 Sep;20(18):1760-1784. doi: 10.1080/15384101.2021.1966584. Epub 2021 Aug 26. Cell Cycle. 2021. PMID: 34437813 Free PMC article. Review.
Cited by
-
Decoding Ubiquitin Modifications by Mass Spectrometry.Adv Exp Med Biol. 2024;1466:1-18. doi: 10.1007/978-981-97-7288-9_1. Adv Exp Med Biol. 2024. PMID: 39546132 Review.
-
Phosphorylation of USP27X by GSK3β maintains the stability and oncogenic functions of CBX2.Cell Death Dis. 2023 Nov 29;14(11):782. doi: 10.1038/s41419-023-06304-y. Cell Death Dis. 2023. PMID: 38030604 Free PMC article.
-
Analysis of a macrophage carbamylated proteome reveals a function in post-translational modification crosstalk.Res Sq [Preprint]. 2023 Jun 16:rs.3.rs-3044777. doi: 10.21203/rs.3.rs-3044777/v1. Res Sq. 2023. Update in: Cell Commun Signal. 2023 Sep 18;21(1):241. doi: 10.1186/s12964-023-01257-3. PMID: 37398265 Free PMC article. Updated. Preprint.
-
The Emerging Role of the Histone H2AK13/15 Ubiquitination: Mechanisms of Writing, Reading, and Erasing in DNA Damage Repair and Disease.Cells. 2025 Feb 18;14(4):307. doi: 10.3390/cells14040307. Cells. 2025. PMID: 39996778 Free PMC article. Review.
-
The ubiquitin codes in cellular stress responses.Protein Cell. 2024 Feb 29;15(3):157-190. doi: 10.1093/procel/pwad045. Protein Cell. 2024. PMID: 37470788 Free PMC article. Review.
References
-
- Ai H, Guo Y, Sun D, Liu S, Qi Y, Guo J, Qu Q, Gong Q, Zhao S, Li J, and Liu L (2019). Examination of the deubiquitylation site selectivity of USP51 by using chemically synthesized ubiquitylated histones. ChemBioChem 20, 221–229. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

