Characterisation and Mutagenesis Study of An Alternative Sigma Factor Gene (hrpL) from Erwinia mallotivora Reveal Its Central Role in Papaya Dieback Disease
- PMID: 33023069
- PMCID: PMC7600996
- DOI: 10.3390/biology9100323
Characterisation and Mutagenesis Study of An Alternative Sigma Factor Gene (hrpL) from Erwinia mallotivora Reveal Its Central Role in Papaya Dieback Disease
Abstract
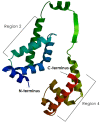
The alternative sigma (σ) factor E, RpoE or HrpL, has been reported to be involved in stress- and pathogenicity-related transcription initiation in Escherichia coli and many other Gram-negative bacteria, including Erwinia spp. and Pseudomonas spp. A previous study identified the hrpL/rpoE transcript as one of the significant differentially expressed genes (DEGs) during early E. mallotivora infection in papaya and those data serve as the basis of the current project. Here, the full coding DNA sequence (CDS) of hrpL from E. mallotivora (EmhrpL) was determined to be 549 bp long, and it encoded a 21.3 kDa HrpL protein that possessed two highly conserved sigma-70 (σ70) motifs-σR2 and σR4. Nucleotide sequence alignment revealed the hrpL from E. mallotivora shared high sequence similarity to rpoE/hrpL from E. tracheiphila (83%), E. pyrifoliae (81%), and E. tasmaniensis (80%). Phylogenetics analysis indicated hrpL from E. mallotivora to be monophyletic with rpoEs/hrpLs from Pantoea vagans, E. herbicola, and E. tracheiphila. Structural analysis postulated that the E. mallotivora's alternative σ factor was non-transmembranic and was an extracytoplasmic function (ECF) protein-characteristics shared by other σ factors in different bacterial species. Notably, the protein-protein interaction (PPI) study through molecular docking suggested the σ factor could be possibly inhibited by an anti-σ. Finally, a knockout of hrpL in E. mallotivora (ΔEmhrpL) resulted in avirulence in four-month-old papaya plants. These findings have revealed that the hrpL is a necessary element in E. mallotivora pathogenicity and also predicted that the gene can be inhibited by an anti-σ.
Keywords: Malaysia; bacterial disease; papaya; transcriptome sequencing.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures







References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

