The murine Microenvironment Cell Population counter method to estimate abundance of tissue-infiltrating immune and stromal cell populations in murine samples using gene expression
- PMID: 33023656
- PMCID: PMC7541325
- DOI: 10.1186/s13073-020-00783-w
The murine Microenvironment Cell Population counter method to estimate abundance of tissue-infiltrating immune and stromal cell populations in murine samples using gene expression
Abstract
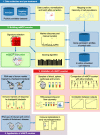
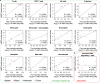
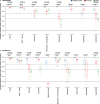
Quantifying tissue-infiltrating immune and stromal cells provides clinically relevant information for various diseases. While numerous methods can quantify immune or stromal cells in human tissue samples from transcriptomic data, few are available for mouse studies. We introduce murine Microenvironment Cell Population counter (mMCP-counter), a method based on highly specific transcriptomic markers that accurately quantify 16 immune and stromal murine cell populations. We validated mMCP-counter with flow cytometry data and showed that mMCP-counter outperforms existing methods. We showed that mMCP-counter scores are predictive of response to immune checkpoint blockade in cancer mouse models and identify early immune impacts of Alzheimer's disease.
Keywords: Alzheimer’s disease; Heterogeneous tissue; Immune checkpoint blockade; Immune composition; Tumor microenvironment.
Conflict of interest statement
WHF is a consultant for Adaptimmune, AstraZeneca, Novartis, Anaveon, Catalym, Oxford Biotherapeutics, OSE immunotherapeutics, Zelluna, and IPSEN. The remaining authors declare that they have no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

