The RNA m6A Reader YTHDF2 Maintains Oncogene Expression and Is a Targetable Dependency in Glioblastoma Stem Cells
- PMID: 33023892
- PMCID: PMC8110214
- DOI: 10.1158/2159-8290.CD-20-0331
The RNA m6A Reader YTHDF2 Maintains Oncogene Expression and Is a Targetable Dependency in Glioblastoma Stem Cells
Abstract
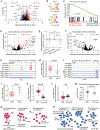
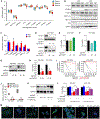
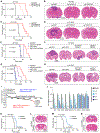
Glioblastoma is a universally lethal cancer driven by glioblastoma stem cells (GSC). Here, we interrogated N 6-methyladenosine (m6A) mRNA modifications in GSCs by methyl RNA immunoprecipitation followed by sequencing and transcriptome analysis, finding transcripts marked by m6A often upregulated compared with normal neural stem cells (NSC). Interrogating m6A regulators, GSCs displayed preferential expression, as well as in vitro and in vivo dependency, of the m6A reader YTHDF2, in contrast to NSCs. Although YTHDF2 has been reported to destabilize mRNAs, YTHDF2 stabilized MYC and VEGFA transcripts in GSCs in an m6A-dependent manner. We identified IGFBP3 as a downstream effector of the YTHDF2-MYC axis in GSCs. The IGF1/IGF1R inhibitor linsitinib preferentially targeted YTHDF2-expressing cells, inhibiting GSC viability without affecting NSCs and impairing in vivo glioblastoma growth. Thus, YTHDF2 links RNA epitranscriptomic modifications and GSC growth, laying the foundation for the YTHDF2-MYC-IGFBP3 axis as a specific and novel therapeutic target in glioblastoma. SIGNIFICANCE: Epitranscriptomics promotes cellular heterogeneity in cancer. RNA m6A landscapes of cancer and NSCs identified cell type-specific dependencies and therapeutic vulnerabilities. The m6A reader YTHDF2 stabilized MYC mRNA specifically in cancer stem cells. Given the challenge of targeting MYC, YTHDF2 presents a therapeutic target to perturb MYC signaling in glioblastoma.This article is highlighted in the In This Issue feature, p. 211.
©2020 American Association for Cancer Research.
Conflict of interest statement
Figures




References
-
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, et al. Identification of a cancer stem cell in human brain tumors. Cancer research 2003;63(18):5821–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA169117/CA/NCI NIH HHS/United States
- R01 GM132292/GM/NIGMS NIH HHS/United States
- R01 CA186702/CA/NCI NIH HHS/United States
- R01 GM110090/GM/NIGMS NIH HHS/United States
- F30 CA217065/CA/NCI NIH HHS/United States
- P30 CA030199/CA/NCI NIH HHS/United States
- R01 CA238662/CA/NCI NIH HHS/United States
- F30 CA217066/CA/NCI NIH HHS/United States
- R01 NS087913/NS/NINDS NIH HHS/United States
- R01 NS103434/NS/NINDS NIH HHS/United States
- R35 CA197718/CA/NCI NIH HHS/United States
- F30 CA203101/CA/NCI NIH HHS/United States
- R01 CA171652/CA/NCI NIH HHS/United States
- R01 NS089272/NS/NINDS NIH HHS/United States
- T32 GM007250/GM/NIGMS NIH HHS/United States
- R01 CA154130/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

