A Novel Mapping Strategy Utilizing Mouse Chromosome Substitution Strains Identifies Multiple Epistatic Interactions That Regulate Complex Traits
- PMID: 33023974
- PMCID: PMC7718749
- DOI: 10.1534/g3.120.401824
A Novel Mapping Strategy Utilizing Mouse Chromosome Substitution Strains Identifies Multiple Epistatic Interactions That Regulate Complex Traits
Abstract
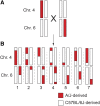
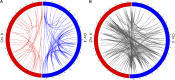
The genetic contribution of additive vs. non-additive (epistatic) effects in the regulation of complex traits is unclear. While genome-wide association studies typically ignore gene-gene interactions, in part because of the lack of statistical power for detecting them, mouse chromosome substitution strains (CSSs) represent an alternate approach for detecting epistasis given their limited allelic variation. Therefore, we utilized CSSs to identify and map both additive and epistatic loci that regulate a range of hematologic- and metabolism-related traits, as well as hepatic gene expression. Quantitative trait loci (QTL) were identified using a CSS-based backcross strategy involving the segregation of variants on the A/J-derived substituted chromosomes 4 and 6 on an otherwise C57BL/6J genetic background. In the liver transcriptomes of offspring from this cross, we identified and mapped additive QTL regulating the hepatic expression of 768 genes, and epistatic QTL pairs for 519 genes. Similarly, we identified additive QTL for fat pad weight, platelets, and the percentage of granulocytes in blood, as well as epistatic QTL pairs controlling the percentage of lymphocytes in blood and red cell distribution width. The variance attributed to the epistatic QTL pairs was approximately equal to that of the additive QTL; however, the SNPs in the epistatic QTL pairs that accounted for the largest variances were undetected in our single locus association analyses. These findings highlight the need to account for epistasis in association studies, and more broadly demonstrate the importance of identifying genetic interactions to understand the complete genetic architecture of complex traits.
Keywords: QTL; complex trait; epistasis; genetic architecture; missing heritability.
Copyright © 2020 Miller et al.
Figures




References
-
- Adler, D. M. D., 2019 rgl: 3D Visualization Using OpenGL. R package version 0.100.30. https://CRAN.R-project.org/package=rgl.
-
- Andrews S., 2010. FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
