End-to-End Protocol for the Detection of SARS-CoV-2 from Built Environments
- PMID: 33024053
- PMCID: PMC7542562
- DOI: 10.1128/mSystems.00771-20
End-to-End Protocol for the Detection of SARS-CoV-2 from Built Environments
Abstract
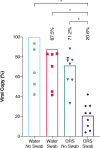
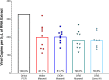
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that causes coronavirus disease 2019, is a respiratory virus primarily transmitted person to person through inhalation of droplets or aerosols, laden with viral particles. However, as recent studies have shown, virions can remain infectious for up to 72 h on surfaces, which can lead to transmission through contact. Thus, a comprehensive study was conducted to determine the efficiency of protocols to recover SARS-CoV-2 from surfaces in built environments. This end-to-end (E2E) study showed that the effective combination for monitoring SARS-CoV-2 on surfaces includes using an Isohelix swab collection tool, DNA/RNA Shield as a preservative, an automated system for RNA extraction, and reverse transcriptase quantitative PCR (RT-qPCR) as the detection assay. Using this E2E approach, this study showed that, in some cases, noninfectious viral fragments of SARS-CoV-2 persisted on surfaces for as long as 8 days even after bleach treatment. Additionally, debris associated with specific built environment surfaces appeared to inhibit and negatively impact the recovery of RNA; Amerstat demonstrated the highest inhibition (>90%) when challenged with an inactivated viral control. Overall, it was determined that this E2E protocol required a minimum of 1,000 viral particles per 25 cm2 to successfully detect virus from test surfaces. Despite our findings of viral fragment longevity on surfaces, when this method was employed to evaluate 368 samples collected from various built environmental surfaces, all samples tested negative, indicating that the surfaces were either void of virus or below the detection limit of the assay.IMPORTANCE The ongoing severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (the virus responsible for coronavirus disease 2019 [COVID-19]) pandemic has led to a global slowdown with far-reaching financial and social impacts. The SARS-CoV-2 respiratory virus is primarily transmitted from person to person through inhalation of infected droplets or aerosols. However, some studies have shown that virions can remain infectious on surfaces for days and can lead to human infection from contact with infected surfaces. Thus, a comprehensive study was conducted to determine the efficiency of protocols to recover SARS-CoV-2 from surfaces in built environments. This end-to-end study showed that the effective combination for monitoring SARS-CoV-2 on surfaces required a minimum of 1,000 viral particles per 25 cm2 to successfully detect virus from surfaces. This comprehensive study can provide valuable information regarding surface monitoring of various materials as well as the capacity to retain viral RNA and allow for effective disinfection.
Keywords: COVID-19; LAMP; RT-qPCR; SARS-CoV-2; built environments; coronavirus; end-to-end; fomites; high-touch surface; surface sampling.
Copyright © 2020 Parker et al.
Figures




References
-
- Ru H, Yang E, Zou K. 2020. What do we learn from SARS-CoV-1 to SARS-CoV-2: evidence from global stock markets. Available at SSRN 3569330. doi:10.2139/ssrn.3569330. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
