SARS-CoV-2 S1 and N-based serological assays reveal rapid seroconversion and induction of specific antibody response in COVID-19 patients
- PMID: 33024213
- PMCID: PMC7538990
- DOI: 10.1038/s41598-020-73491-5
SARS-CoV-2 S1 and N-based serological assays reveal rapid seroconversion and induction of specific antibody response in COVID-19 patients
Abstract
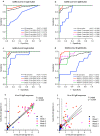
As the Coronavirus Disease 2019 (COVID-19), which is caused by the novel SARS-CoV-2, continues to spread rapidly around the world, there is a need for well validated serological assays that allow the detection of viral specific antibody responses in COVID-19 patients or recovered individuals. In this study, we established and used multiple indirect Enzyme Linked Immunosorbent Assay (ELISA)-based serological assays to study the antibody response in COVID-19 patients. In order to validate the assays we determined the cut off values, sensitivity and specificity of the assays using sera collected from pre-pandemic healthy controls, COVID-19 patients at different time points after disease-onset, and seropositive sera to other human coronaviruses (CoVs). The developed SARS-CoV-2 S1 subunit of the spike glycoprotein and nucleocapsid (N)-based ELISAs not only showed high specificity and sensitivity but also did not show any cross-reactivity with other CoVs. We also show that all RT-PCR confirmed COVID-19 patients tested in our study developed both virus specific IgM and IgG antibodies as early as week one after disease onset. Our data also suggest that the inclusion of both S1 and N in serological testing would capture as many potential SARS-CoV-2 positive cases as possible than using any of them alone. This is specifically important for tracing contacts and cases and conducting large-scale epidemiological studies to understand the true extent of virus spread in populations.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Fact Sheet for Health Care Providers Emergency Use Authorization (EUA) of Remdesivir (GS-5734). The U.S. Food and Drug Administration (FDA)https://www.fda.gov/media/137566/download (2020).
-
- Russian Ministry of Health approves the first COVID-19 drug Avifavir produced by JV of RDIF and ChemRar. Russian Direct Investment Fundhttps://rdif.ru/Eng_fullNews/5220/ (2020).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

