Analysis of dynamic and widespread lncRNA and miRNA expression in fetal sheep skeletal muscle
- PMID: 33024632
- PMCID: PMC7518186
- DOI: 10.7717/peerj.9957
Analysis of dynamic and widespread lncRNA and miRNA expression in fetal sheep skeletal muscle
Abstract
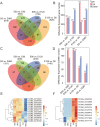
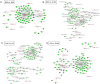
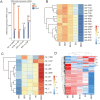
The sheep is an economically important animal, and there is currently a major focus on improving its meat quality through breeding. There are variations in the growth regulation mechanisms of different sheep breeds, making fundamental research on skeletal muscle growth essential in understanding the regulation of (thus far) unknown genes. Skeletal muscle development is a complex biological process regulated by numerous genes and non-coding RNAs, including microRNAs (miRNAs) and long non-coding RNAs (lncRNAs). In this study, we used deep sequencing data from sheep longissimus dorsi (LD) muscles sampled at day 60, 90, and 120 of gestation, as well as at day 0 and 360 following birth, to identify and examine the lncRNA and miRNA temporal expression profiles that regulate sheep skeletal myogenesis. We stained LD muscles using histological sections to analyse the area and circumference of muscle fibers from the embryonic to postnatal development stages. Our results showed that embryonic skeletal muscle growth can be characterized by time. We obtained a total of 694 different lncRNAs and compared the differential expression between the E60 vs. E90, E90 vs. E120, E120 vs. D0, and D0 vs. D360 lncRNA and gene samples. Of the total 701 known sheep miRNAs we detected, the following showed a wide range of expression during the embryonic stage: miR-2387, miR-105, miR-767, miR-432, and miR-433. We propose that the detected lncRNA expression was time-specific during the gestational and postnatal stages. GO and KEGG analyses of the genes targeted by different miRNAs and lncRNAs revealed that these significantly enriched processes and pathways were consistent with skeletal muscle development over time across all sampled stages. We found four visual lncRNA-gene regulatory networks that can be used to explore the function of lncRNAs in sheep and may be valuable in helping improve muscle growth. This study also describes the function of several lncRNAs that interact with miRNAs to regulate myogenic differentiation.
Keywords: Transcriptome; Fetal sheep; Longissimus dorsi muscle; lncRNA; miRNA.
©2020 Yuan et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures


Similar articles
-
Identification and Expression Profiling of miRNAome in Goat longissimus dorsi Muscle from Prenatal Stages to a Neonatal Stage.PLoS One. 2016 Oct 31;11(10):e0165764. doi: 10.1371/journal.pone.0165764. eCollection 2016. PLoS One. 2016. PMID: 27798673 Free PMC article.
-
Integrated analysis of the whole transcriptome of skeletal muscle reveals the ceRNA regulatory network related to the formation of muscle fibers in Tan sheep.Front Genet. 2022 Oct 18;13:991606. doi: 10.3389/fgene.2022.991606. eCollection 2022. Front Genet. 2022. PMID: 36330447 Free PMC article.
-
MicroRNA expression profiles differ between primary myofiber of lean and obese pig breeds.PLoS One. 2017 Jul 31;12(7):e0181897. doi: 10.1371/journal.pone.0181897. eCollection 2017. PLoS One. 2017. PMID: 28759650 Free PMC article.
-
[Regulatory mechanism for lncRNAs in skeletal muscle development and progress on its research in domestic animals].Yi Chuan. 2018 Apr 20;40(4):292-304. doi: 10.16288/j.yczz.17-358. Yi Chuan. 2018. PMID: 29704375 Review. Chinese.
-
Functions and Regulatory Mechanisms of lncRNAs in Skeletal Myogenesis, Muscle Disease and Meat Production.Cells. 2019 Sep 19;8(9):1107. doi: 10.3390/cells8091107. Cells. 2019. PMID: 31546877 Free PMC article. Review.
Cited by
-
MiR-22-3p Inhibits Proliferation and Promotes Differentiation of Skeletal Muscle Cells by Targeting IGFBP3 in Hu Sheep.Animals (Basel). 2022 Jan 4;12(1):114. doi: 10.3390/ani12010114. Animals (Basel). 2022. PMID: 35011220 Free PMC article.
-
Integrated analysis of mRNAs and lncRNAs reveals candidate marker genes and potential hub lncRNAs associated with growth regulation of the Pacific Oyster, Crassostrea gigas.BMC Genomics. 2023 Aug 10;24(1):453. doi: 10.1186/s12864-023-09543-7. BMC Genomics. 2023. PMID: 37563567 Free PMC article.
-
RNA-Seq Reveals miRNA and mRNA Co-regulate Muscle Differentiation in Fetal Leizhou Goats.Front Vet Sci. 2022 Mar 25;9:829769. doi: 10.3389/fvets.2022.829769. eCollection 2022. Front Vet Sci. 2022. PMID: 35400087 Free PMC article.
-
Identification and Characterization of lncRNAs Expression Profile Related to Goat Skeletal Muscle at Different Development Stages.Animals (Basel). 2022 Oct 6;12(19):2683. doi: 10.3390/ani12192683. Animals (Basel). 2022. PMID: 36230427 Free PMC article.
-
Deep Small RNA Sequencing Reveals Important miRNAs Related to Muscle Development and Intramuscular Fat Deposition in Longissimus dorsi Muscle From Different Goat Breeds.Front Vet Sci. 2022 Jun 13;9:911166. doi: 10.3389/fvets.2022.911166. eCollection 2022. Front Vet Sci. 2022. PMID: 35769318 Free PMC article.
References
-
- Auber L. VII—the anatomy of follicles producing wool-fibres, with special reference to Keratinization. Earth and Environmental Science Transactions of the Royal Society of Edinburgh. 1952;62:191–254. doi: 10.1017/S0080456800009285. - DOI
-
- Carter H, Clarke W. Hair follicle group and skin follicle population of Australian Merino sheep. Australian Journal of Agricultural Research. 1957;8:91–108. doi: 10.1071/AR9570091. - DOI
LinkOut - more resources
Full Text Sources
Research Materials

