This is a preprint.
Molecular Architecture of Early Dissemination and Massive Second Wave of the SARS-CoV-2 Virus in a Major Metropolitan Area
- PMID: 33024977
- PMCID: PMC7536878
- DOI: 10.1101/2020.09.22.20199125
Molecular Architecture of Early Dissemination and Massive Second Wave of the SARS-CoV-2 Virus in a Major Metropolitan Area
Update in
-
Molecular Architecture of Early Dissemination and Massive Second Wave of the SARS-CoV-2 Virus in a Major Metropolitan Area.mBio. 2020 Oct 30;11(6):e02707-20. doi: 10.1128/mBio.02707-20. mBio. 2020. PMID: 33127862 Free PMC article.
Abstract
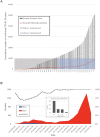
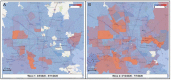
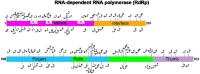
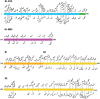
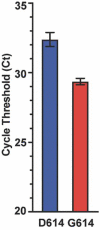
We sequenced the genomes of 5,085 SARS-CoV-2 strains causing two COVID-19 disease waves in metropolitan Houston, Texas, an ethnically diverse region with seven million residents. The genomes were from viruses recovered in the earliest recognized phase of the pandemic in Houston, and an ongoing massive second wave of infections. The virus was originally introduced into Houston many times independently. Virtually all strains in the second wave have a Gly614 amino acid replacement in the spike protein, a polymorphism that has been linked to increased transmission and infectivity. Patients infected with the Gly614 variant strains had significantly higher virus loads in the nasopharynx on initial diagnosis. We found little evidence of a significant relationship between virus genotypes and altered virulence, stressing the linkage between disease severity, underlying medical conditions, and host genetics. Some regions of the spike protein - the primary target of global vaccine efforts - are replete with amino acid replacements, perhaps indicating the action of selection. We exploited the genomic data to generate defined single amino acid replacements in the receptor binding domain of spike protein that, importantly, produced decreased recognition by the neutralizing monoclonal antibody CR30022. Our study is the first analysis of the molecular architecture of SARS-CoV-2 in two infection waves in a major metropolitan region. The findings will help us to understand the origin, composition, and trajectory of future infection waves, and the potential effect of the host immune response and therapeutic maneuvers on SARS-CoV-2 evolution.
Figures


References
-
- 2020. World Health Organization Coronavirus Disease 2019 (COVID-19) Situation Report. https://www.who.int/docs/default-source/coronaviruse/situation-reports/2.... Accessed April 21.
-
- Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA, Haagmans BL, Lauber C, Leontovich AM, Neuman BW, Penzar D, Perlman S, Poon LLM, Samborskiy DV, Sidorov IA, Sola I, Ziebuhr J, Coronaviridae Study Group of the International Committee on Taxonomy of V. 2020. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiology 5:536–544. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
