This is a preprint.
Viral genomes reveal patterns of the SARS-CoV-2 outbreak in Washington State
- PMID: 33024981
- PMCID: PMC7536883
- DOI: 10.1101/2020.09.30.20204230
Viral genomes reveal patterns of the SARS-CoV-2 outbreak in Washington State
Update in
-
Viral genomes reveal patterns of the SARS-CoV-2 outbreak in Washington State.Sci Transl Med. 2021 May 26;13(595):eabf0202. doi: 10.1126/scitranslmed.abf0202. Epub 2021 May 3. Sci Transl Med. 2021. PMID: 33941621 Free PMC article.
Abstract
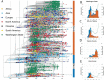
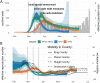
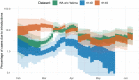
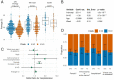
The rapid spread of SARS-CoV-2 has gravely impacted societies around the world. Outbreaks in different parts of the globe are shaped by repeated introductions of new lineages and subsequent local transmission of those lineages. Here, we sequenced 3940 SARS-CoV-2 viral genomes from Washington State to characterize how the spread of SARS-CoV-2 in Washington State (USA) was shaped by differences in timing of mitigation strategies across counties, as well as by repeated introductions of viral lineages into the state. Additionally, we show that the increase in frequency of a potentially more transmissible viral variant (614G) over time can potentially be explained by regional mobility differences and multiple introductions of 614G, but not the other variant (614D) into the state. At an individual level, we see evidence of higher viral loads in patients infected with the 614G variant. However, using clinical records data, we do not find any evidence that the 614G variant impacts clinical severity or patient outcomes. Overall, this suggests that at least to date, the behavior of individuals has been more important in shaping the course of the pandemic than changes in the virus.
Conflict of interest statement
Competing interests
Janet A. Englund is a consultant for Sanofi Pasteur and Meissa Vaccines, Inc., and receives research support from GlaxoSmithKline, AstraZeneca, and Novavax. Helen Chu is a consultant for Merck and GlaxoSmithKline. Jay Shendure is a consultant with Guardant Health, Maze Therapeutics, Camp4 Therapeutics, Nanostring, Phase Genomics, Adaptive Biotechnologies, and Stratos Genomics, and has a research collaboration with Illumina. Michael Boeckh is a consultant for Merck and VirBio. Nicola F. Müller, Cassia Wagner, Chris D. Frazar, Pavitra Roychoudhury, Jover Lee, Louise H. Moncla, Benjamin Pelle, Matthew Richardson, Erica Ryke, Hong Xie, Lasata Shrestha, Amin Addetia, Victoria M. Rachleff, Nicole A. P. Lieberman, Meei-Li Huang, Romesh Gautom, Geoff Melly, Brian Hiatt, Philip Dykema, Amanda Adler, Elisabeth Brandstetter, Peter D. Han, Kairsten Fay, Misja Ilcisin, Kirsten Lacombe, Thomas R. Sibley, Melissa Truong, Caitlin R. Wolf, Michael Famulare, Barry R. Lutz, Mark J. Rieder, Matthew Thompson, Jeffrey S. Duchin, Lea M. Starita, Keith R. Jerome, Scott Lindquist, Alexander L. Greninger, Deborah A. Nickerson, and Trevor Bedford declare no competing interests.
Figures
References
-
- Rambaut A, Phylogenetic analysis of nCoV-2019 genomes Virological (available at http://virological.org/t/phylodynamic-analysis-176-genomes-6-mar-2020/356).
-
- Bedford T., Greninger A. L., Roychoudhury P., Starita L. M., Famulare M., Huang M.-L., Nalla A., Pepper G., Reinhardt A., Xie H., Shrestha L., Nguyen T. N., Adler A., Brandstetter E., Cho S., Giroux D., Han P. D., Fay K., Frazar C. D., Ilcisin M., Lacombe K., Lee J., Kiavand A., Richardson M., Sibley T. R., Truong M., Wolf C. R., Nickerson D. A., Rieder M. J., Englund J. A., Hadfield J., Hodcroft E. B., Huddleston J., Moncla L. H., Müller N. F., Neher R. A., Deng X., Gu W., Federman S., Chiu C., Duchin J., Gautom R., Melly G., Hiatt B., Dykema P., Lindquist S., Queen K., Tao Y., Uehara A., Tong S., MacCannell D., Armstrong G. L., Baird G. S., Chu H. Y., Shendure J., Jerome K. R., Cryptic transmission of SARS-CoV-2 in Washington State, Science, eabc0523 (2020). - PMC - PubMed
-
- Korber B., Fischer W. M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E. E., Bhattacharya T., Foley B., Hastie K. M., Parker M. D., Partridge D. G., Evans C. M., Freeman T. M., de Silva T. I., McDanal C., Perez L. G., Tang H., Moon-Walker A., Whelan S. P., LaBranche C. C., Saphire E. O., Montefiori D. C., Angyal A., Brown R. L., Carrilero L., Green L. R., Groves D. C., Johnson K. J., Keeley A. J., Lindsey B. B., Parsons P. J., Raza M., Rowland-Jones S., Smith N., Tucker R. M., Wang D., Wyles M. D., Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus Cell (2020), doi: 10.1016/j.cell.2020.06.043. - DOI - PMC - PubMed
-
- Yurkovetskiy L., Pascal K. E., Tompkins-Tinch C., Nyalile T., Wang Y., Baum A., Diehl W. E., Dauphin A., Carbone C., Veinotte K., Egri S. B., Schaffner S. F., Lemieux J. E., Munro J., Sabeti P. C., Kyratsous C., Shen K., Luban J., SARS-CoV-2 Spike protein variant D614G increases infectivity and retains sensitivity to antibodies that target the receptor binding domain, bioRxiv (2020), doi: 10.1016/j.cell.2020.09.032 - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
