Epigenomic, genomic, and transcriptomic landscape of schwannomatosis
- PMID: 33025139
- PMCID: PMC7785562
- DOI: 10.1007/s00401-020-02230-x
Epigenomic, genomic, and transcriptomic landscape of schwannomatosis
Erratum in
-
Correction to: Epigenomic, genomic, and transcriptomic landscape of schwannomatosis.Acta Neuropathol. 2021 Jan;141(1):117. doi: 10.1007/s00401-020-02241-8. Acta Neuropathol. 2021. PMID: 33112994 Free PMC article. No abstract available.
Abstract
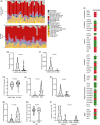
Schwannomatosis (SWNTS) is a genetic cancer predisposition syndrome that manifests as multiple and often painful neuronal tumors called schwannomas (SWNs). While germline mutations in SMARCB1 or LZTR1, plus somatic mutations in NF2 and loss of heterozygosity in chromosome 22q have been identified in a subset of patients, little is known about the epigenomic and genomic alterations that drive SWNTS-related SWNs (SWNTS-SWNs) in a majority of the cases. We performed multiplatform genomic analysis and established the molecular signature of SWNTS-SWNs. We show that SWNTS-SWNs harbor distinct genomic features relative to the histologically identical non-syndromic sporadic SWNs (NS-SWNS). We demonstrate the existence of four distinct DNA methylation subgroups of SWNTS-SWNs that are associated with specific transcriptional programs and tumor location. We show several novel recurrent non-22q deletions and structural rearrangements. We detected the SH3PXD2A-HTRA1 gene fusion in SWNTS-SWNs, with predominance in LZTR1-mutant tumors. In addition, we identified specific genetic, epigenetic, and actionable transcriptional programs associated with painful SWNTS-SWNs including PIGF, VEGF, MEK, and MTOR pathways, which may be harnessed for management of this syndrome.
Keywords: Genomics; LZTR1; MAPK; Pain; Peripheral nerve sheath tumors; Schwannomatosis.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

