Compartment-specific transcriptomics of ozone-exposed murine lungs reveals sex- and cell type-associated perturbations relevant to mucoinflammatory lung diseases
- PMID: 33026818
- PMCID: PMC7847060
- DOI: 10.1152/ajplung.00381.2020
Compartment-specific transcriptomics of ozone-exposed murine lungs reveals sex- and cell type-associated perturbations relevant to mucoinflammatory lung diseases
Abstract
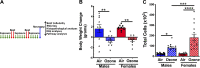
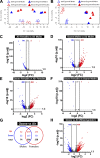
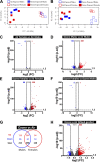
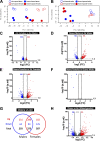
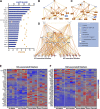
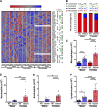
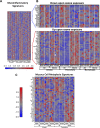
Ozone is known to cause lung injury, and resident cells of the respiratory tract (i.e., epithelial cells and macrophages) respond to inhaled ozone in a variety of ways that affect their survival, morphology, and functioning. However, a complete understanding of the sex-associated and the cell type-specific gene expression changes in response to ozone exposure is still limited. Through transcriptome profiling, we aimed to analyze gene expression alterations and associated enrichment of biological pathways in three distinct cell type-enriched compartments of ozone-exposed murine lungs. We subchronically exposed adult male and female mice to 0.8 ppm ozone or filtered air. RNA-Seq was performed on airway epithelium-enriched airways, parenchyma, and purified airspace macrophages. Differential gene expression and biological pathway analyses were performed and supported by cellular and immunohistochemical analyses. While a majority of differentially expressed genes (DEGs) in ozone-exposed versus air-exposed groups were common between both sexes, sex-specific DEGs were also identified in all of the three tissue compartments. As compared with ozone-exposed males, ozone-exposed females had significant alterations in gene expression in three compartments. Pathways relevant to cell division and DNA repair were enriched in the ozone-exposed airways, indicating ozone-induced airway injury and repair, which was further supported by immunohistochemical analyses. In addition to cell division and DNA repair pathways, inflammatory pathways were also enriched within the parenchyma, supporting contribution by both epithelial and immune cells. Further, immune response and cytokine-cytokine receptor interactions were enriched in macrophages, indicating ozone-induced macrophage activation. Finally, our analyses also revealed the overall upregulation of mucoinflammation- and mucous cell metaplasia-associated pathways following ozone exposure.
Keywords: airways; gene expression; macrophages; ozone; parenchyma.
Conflict of interest statement
No conflicts of interest, financial or otherwise, are declared by the authors.
Figures





References
-
- Anenberg SC, Henze DK, Tinney V, Kinney PL, Raich W, Fann N, Malley CS, Roman H, Lamsal L, Duncan B, Martin RV, van Donkelaar A, Brauer M, Doherty R, Jonson JE, Davila Y, Sudo K, Kuylenstierna JCI. Estimates of the global burden of ambient PM2.5, ozone, and NO2 on asthma incidence and emergency room visits. Environ Health Perspect 126: 107004, 2018. doi:10.1289/EHP3766. - DOI - PMC - PubMed
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G; The Gene Ontology Consortium . Gene ontology: tool for the unification of biology. Nat Genet 25: 25–29, 2000. doi:10.1038/75556. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

