LipidFinder 2.0: advanced informatics pipeline for lipidomics discovery applications
- PMID: 33027502
- PMCID: PMC8208733
- DOI: 10.1093/bioinformatics/btaa856
LipidFinder 2.0: advanced informatics pipeline for lipidomics discovery applications
Abstract
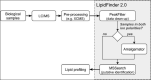
Summary: We present LipidFinder 2.0, incorporating four new modules that apply artefact filters, remove lipid and contaminant stacks, in-source fragments and salt clusters, and a new isotope deletion method which is significantly more sensitive than available open-access alternatives. We also incorporate a novel false discovery rate method, utilizing a target-decoy strategy, which allows users to assess data quality. A renewed lipid profiling method is introduced which searches three different databases from LIPID MAPS and returns bulk lipid structures only, and a lipid category scatter plot with color blind friendly pallet. An API interface with XCMS Online is made available on LipidFinder's online version. We show using real data that LipidFinder 2.0 provides a significant improvement over non-lipid metabolite filtering and lipid profiling, compared to available tools.
Availability and implementation: LipidFinder 2.0 is freely available at https://github.com/ODonnell-Lipidomics/LipidFinder and http://lipidmaps.org/resources/tools/lipidfinder.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2020. Published by Oxford University Press.
Figures
References
-
- Liebisch G. et al. (2015) Identification and annotation of lipid species in metabolomics studies need improvement. Clin. Chem., 61, 1542–1544. - PubMed