Sox9 deletion causes severe intervertebral disc degeneration characterized by apoptosis, matrix remodeling, and compartment-specific transcriptomic changes
- PMID: 33027692
- PMCID: PMC7778523
- DOI: 10.1016/j.matbio.2020.09.003
Sox9 deletion causes severe intervertebral disc degeneration characterized by apoptosis, matrix remodeling, and compartment-specific transcriptomic changes
Abstract
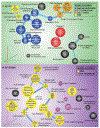
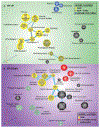
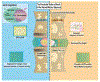
SOX9 plays an important role in chondrocyte differentiation and, in the developing axial skeleton, maintains the notochord and the demarcation of intervertebral disc compartments. Diminished expression is linked to campomelic dysplasia, resulting in severe scoliosis and progressive disc degeneration. However, the specific functions of SOX9 in the adult spinal column and disc are largely unknown. Accordingly, employing a strategy to conditionally delete Sox9 in Acan-expressing cells (AcanCreERT2Sox9fl/fl), we delineated these functions in the adult intervertebral disc. AcanCreERT2Sox9fl/fl mice (Sox9cKO) showed extensive and progressive remodeling of the extracellular matrix in nucleus pulposus (NP) and annulus fibrosus (AF), consistent with human disc degeneration. Progressive degeneration of the cartilaginous endplates (EP) was also evident in Sox9cKO mice, and it preceded morphological changes seen in the NP and AF compartments. Fate mapping using tdTomato reporter, EdU chase, and quantitative immunohistological studies demonstrated that SOX9 is crucial for disc cell survival and phenotype maintenance. Microarray analysis showed that Sox9 regulated distinct compartment-specific transcriptomic landscapes, with prominent contributions to the ECM, cytoskeleton-related, and metabolic pathways in the NP and ion transport, the cell cycle, and signaling pathways in the AF. In summary, our work provides new insights into disc degeneration in Sox9cKO mice at the cellular, molecular, and transcriptional levels, underscoring tissue-specific roles of this transcription factor. Our findings may direct future cell therapies targeting SOX9 to mitigate disc degeneration.
Keywords: Annulus fibrosus; Extracellular matrix; Intervertebral disc degeneration; Mouse model; Nucleus pulposus; Sox9.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest R.A. Barve may receive royalty income based on the CompBio technology developed by R.A. Barve and licensed by Washington University to PercayAI. The remaining authors declare they have no competing interests to disclose in relation to the contents of this article.
Figures







References
-
- Global Burden of Disease Study 2013 Collaborators (2015). Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 386, 743–800. doi: 10.1016/S0140-6736(15)60692-4 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

