Quantification of low affinity binding interactions between natural killer cell inhibitory receptors and targeting ligands with a self-induced back-action actuated nanopore electrophoresis (SANE) sensor
- PMID: 33027774
- PMCID: PMC8346883
- DOI: 10.1088/1361-6528/abbf26
Quantification of low affinity binding interactions between natural killer cell inhibitory receptors and targeting ligands with a self-induced back-action actuated nanopore electrophoresis (SANE) sensor
Abstract
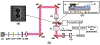
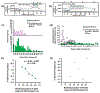
A plasmonic nanopore sensor enabling detection of bimodal optical and electrical molecular signatures was fabricated and tested for its ability to characterize low affinity ligand-receptor interactions. This plasmonic nanosensor uses self-induced back-action (SIBA) for optical trapping to enable SIBA-actuated nanopore electrophoresis (SANE) through a nanopore located immediately below the optical trap volume. A natural killer (NK) cell inhibitory receptor heterodimer molecule CD94/NKG2A was synthesized to target a specific peptide-presenting Qa-1b Qdm ligand as a simplified model of low-affinity interactions between immune cells and peptide-presenting cancer cells that occurs during cancer immunotherapy. A cancer-irrelevant Qa-1b GroEL ligand was also targeted by the same receptor as a control experiment to test for non-specific binding. The analysis of different pairs of bimodal SANE sensor signatures enabled discrimination of ligand, receptor and their complexes and enabled differentiating between specific and non-specific ligand interactions. We were able to detect ligand-receptor complex binding at concentrations over 500 times lower than the free solution equilibrium binding constant (K D ). Additionally, SANE sensor measurements enabled estimation of the fast dissociation rate (k off) for this low-affinity specific ligand-receptor system, previously shown to be challenging to quantify with commercial technologies. The k off value of targeted peptide-presenting ligands is known to correlate with the subsequent activation of immune cells in vivo, suggesting the potential utility of the SANE senor as a screening tool in cancer immunotherapy.
Figures









References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
