A comparative analysis of chromatin accessibility in cattle, pig, and mouse tissues
- PMID: 33028202
- PMCID: PMC7541309
- DOI: 10.1186/s12864-020-07078-9
A comparative analysis of chromatin accessibility in cattle, pig, and mouse tissues
Abstract
Background: Although considerable progress has been made towards annotating the noncoding portion of the human and mouse genomes, regulatory elements in other species, such as livestock, remain poorly characterized. This lack of functional annotation poses a substantial roadblock to agricultural research and diminishes the value of these species as model organisms. As active regulatory elements are typically characterized by chromatin accessibility, we implemented the Assay for Transposase Accessible Chromatin (ATAC-seq) to annotate and characterize regulatory elements in pigs and cattle, given a set of eight adult tissues.
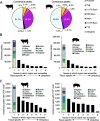
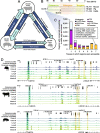
Results: Overall, 306,304 and 273,594 active regulatory elements were identified in pig and cattle, respectively. 71,478 porcine and 47,454 bovine regulatory elements were highly tissue-specific and were correspondingly enriched for binding motifs of known tissue-specific transcription factors. However, in every tissue the most prevalent accessible motif corresponded to the insulator CTCF, suggesting pervasive involvement in 3-D chromatin organization. Taking advantage of a similar dataset in mouse, open chromatin in pig, cattle, and mice were compared, revealing that the conservation of regulatory elements, in terms of sequence identity and accessibility, was consistent with evolutionary distance; whereas pig and cattle shared about 20% of accessible sites, mice and ungulates only had about 10% of accessible sites in common. Furthermore, conservation of accessibility was more prevalent at promoters than at intergenic regions.
Conclusions: The lack of conserved accessibility at distal elements is consistent with rapid evolution of enhancers, and further emphasizes the need to annotate regulatory elements in individual species, rather than inferring elements based on homology. This atlas of chromatin accessibility in cattle and pig constitutes a substantial step towards annotating livestock genomes and dissecting the regulatory link between genome and phenome.
Keywords: Cattle; Chromatin accessibility; Comparative epigenomics; Functional annotation; Pig.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

