Symbiotic microbiome Staphylococcus aureus from human nasal mucus modulates IL-33-mediated type 2 immune responses in allergic nasal mucosa
- PMID: 33028252
- PMCID: PMC7542126
- DOI: 10.1186/s12866-020-01974-6
Symbiotic microbiome Staphylococcus aureus from human nasal mucus modulates IL-33-mediated type 2 immune responses in allergic nasal mucosa
Abstract
Background: The host-microbial commensalism can shape the innate immune responses in respiratory mucosa and nasal microbiome also modulates front-line immune mechanism in the nasal mucosa. Inhaled allergens encounter the host immune system first in the nasal mucosa, and microbial characteristics of nasal mucus directly impact the mechanisms of initial allergic responses in nasal epithelium. However, the roles of the nasal microbiome in allergic nasal mucosa remain uncertain. We sought to determine the distribution of nasal microbiomes in allergic nasal mucosa and elucidate the interplay between nasal microbiome Staphylococcus species and Th2 cytokines in allergic rhinitis (AR) models.
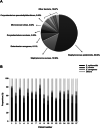
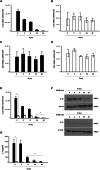
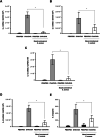
Results: Staphylococcus aureus (AR-SA) and S. epidermidis (AR-SE) were isolated from the nasal mucosa of patients with AR. The influence of nasal microbiome Staphylococcus species on allergic nasal mucosa was also tested with in vitro and in vivo AR models. Pyrosequencing data showed that colonization by S. epidermidis and S. aureus was more dominant in nasal mucus of AR subjects. The mRNA and protein levels of IL-33 and TSLP were significantly higher in AR nasal epithelial (ARNE) cells which were cultured from nasal mucosa of AR subjects, and exposure of ARNE cells to AR-SA reduced IL-33 mRNA and secreted protein levels. Particularly, ovalbumin-driven AR mice inoculated with AR-SA by intranasal delivery exhibited significantly reduced IL-33 in their nasal mucosa. In the context of these results, allergic symptoms and Th2 cytokine levels were significantly downregulated after intranasal inoculation of AR-SA in vivo AR mice.
Conclusion: Colonization by Staphylococcus species was more dominant in allergic nasal mucosa, and nasal commensal S. aureus from subjects with AR mediates anti-allergic effects by modulating IL-33-dependent Th2 inflammation. The results demonstrate the role of host-bacterial commensalism in shaping human allergic inflammation.
Keywords: Allergic rhinitis; Interleukin-33; Nasal microbiome; Staphylococcus aureus; Symbiosis.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Figures


References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Research Materials

