A sleep-like state in Hydra unravels conserved sleep mechanisms during the evolutionary development of the central nervous system
- PMID: 33028524
- PMCID: PMC7541080
- DOI: 10.1126/sciadv.abb9415
A sleep-like state in Hydra unravels conserved sleep mechanisms during the evolutionary development of the central nervous system
Abstract
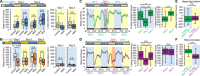
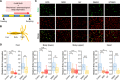
Sleep behaviors are observed even in nematodes and arthropods, yet little is known about how sleep-regulatory mechanisms have emerged during evolution. Here, we report a sleep-like state in the cnidarian Hydra vulgaris with a primitive nervous organization. Hydra sleep was shaped by homeostasis and necessary for cell proliferation, but it lacked free-running circadian rhythms. Instead, we detected 4-hour rhythms that might be generated by ultradian oscillators underlying Hydra sleep. Microarray analysis in sleep-deprived Hydra revealed sleep-dependent expression of 212 genes, including cGMP-dependent protein kinase 1 (PRKG1) and ornithine aminotransferase. Sleep-promoting effects of melatonin, GABA, and PRKG1 were conserved in Hydra However, arousing dopamine unexpectedly induced Hydra sleep. Opposing effects of ornithine metabolism on sleep were also evident between Hydra and Drosophila, suggesting the evolutionary switch of their sleep-regulatory functions. Thus, sleep-relevant physiology and sleep-regulatory components may have already been acquired at molecular levels in a brain-less metazoan phylum and reprogrammed accordingly.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures




References
-
- Anafi R. C., Kayser M. S., Raizen D. M., Exploring phylogeny to find the function of sleep. Nat. Rev. Neurosci. 20, 109–116 (2019). - PubMed
-
- Garm A., Bielecki J., Petie R., Nilsson D.-E., Opposite patterns of diurnal activity in the box jellyfish Tripedalia cystophora and Copula sivickisi. Biol. Bull. 222, 35–45 (2012). - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

