Analysis of the molecular nature associated with microsatellite status in colon cancer identifies clinical implications for immunotherapy
- PMID: 33028695
- PMCID: PMC7542666
- DOI: 10.1136/jitc-2020-001437
Analysis of the molecular nature associated with microsatellite status in colon cancer identifies clinical implications for immunotherapy
Abstract
Background: Microsatellite instability in colon cancer implies favorable therapeutic outcomes after checkpoint blockade immunotherapy. However, the molecular nature of microsatellite instability is not well elucidated.
Methods: We examined the immune microenvironment of colon cancer using assessments of the bulk transcriptome and the single-cell transcriptome focusing on molecular nature of microsatellite stability (MSS) and microsatellite instability (MSI) in colorectal cancer from a public database. The association of the mutation pattern and microsatellite status was analyzed by a random forest algorithm in The Cancer Genome Atlas (TCGA) and validated by our in-house dataset (39 tumor mutational burden (TMB)-low MSS colon cancer, 10 TMB-high MSS colon cancer, 15 MSI colon cancer). A prognostic model was constructed to predict the survival potential and stratify microsatellite status by a neural network.
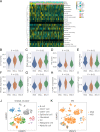
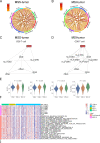
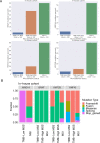
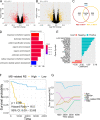
Results: Despite the hostile CD8+ cytotoxic T lymphocyte (CTL)/Th1 microenvironment in MSI colon cancer, a high percentage of exhausted CD8+ T cells and upregulated expression of immune checkpoints were identified in MSI colon cancer at the single-cell level, indicating the potential neutralizing effect of cytotoxic T-cell activity by exhausted T-cell status. A more homogeneous highly expressed pattern of PD1 was observed in CD8+ T cells from MSI colon cancer; however, a small subgroup of CD8+ T cells with high expression of checkpoint molecules was identified in MSS patients. A random forest algorithm predicted important mutations that were associated with MSI status in the TCGA colon cancer cohort, and our in-house cohort validated higher frequencies of BRAF, ARID1A, RNF43, and KM2B mutations in MSI colon cancer. A robust microsatellite status-related gene signature was built to predict the prognosis and differentiate between MSI and MSS tumors. A neural network using the expression profile of the microsatellite status-related gene signature was constructed. A receiver operating characteristic curve was used to evaluate the accuracy rate of neural network, reaching 100%.
Conclusion: Our analysis unraveled the difference in the molecular nature and genomic variance in MSI and MSS colon cancer. The microsatellite status-related gene signature is better at predicting the prognosis of patients with colon cancer and response to the combination of immune checkpoint inhibitor-based immunotherapy and anti-VEGF therapy.
Keywords: computational biology; immunotherapy; tumor biomarkers; tumor microenvironment.
© Author(s) (or their employer(s)) 2020. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
