Overexpression of Cyclin E1 or Cdc25A leads to replication stress, mitotic aberrancies, and increased sensitivity to replication checkpoint inhibitors
- PMID: 33028815
- PMCID: PMC7542455
- DOI: 10.1038/s41389-020-00270-2
Overexpression of Cyclin E1 or Cdc25A leads to replication stress, mitotic aberrancies, and increased sensitivity to replication checkpoint inhibitors
Abstract
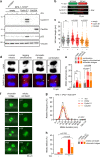
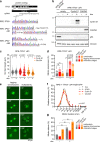
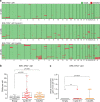
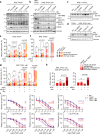
Oncogene-induced replication stress, for instance as a result of Cyclin E1 overexpression, causes genomic instability and has been linked to tumorigenesis. To survive high levels of replication stress, tumors depend on pathways to deal with these DNA lesions, which represent a therapeutically actionable vulnerability. We aimed to uncover the consequences of Cyclin E1 or Cdc25A overexpression on replication kinetics, mitotic progression, and the sensitivity to inhibitors of the WEE1 and ATR replication checkpoint kinases. We modeled oncogene-induced replication stress using inducible expression of Cyclin E1 or Cdc25A in non-transformed RPE-1 cells, either in a TP53 wild-type or TP53-mutant background. DNA fiber analysis showed Cyclin E1 or Cdc25A overexpression to slow replication speed. The resulting replication-derived DNA lesions were transmitted into mitosis causing chromosome segregation defects. Single cell sequencing revealed that replication stress and mitotic defects upon Cyclin E1 or Cdc25A overexpression resulted in genomic instability. ATR or WEE1 inhibition exacerbated the mitotic aberrancies induced by Cyclin E1 or Cdc25A overexpression, and caused cytotoxicity. Both these phenotypes were exacerbated upon p53 inactivation. Conversely, downregulation of Cyclin E1 rescued both replication kinetics, as well as sensitivity to ATR and WEE1 inhibitors. Taken together, Cyclin E1 or Cdc25A-induced replication stress leads to mitotic segregation defects and genomic instability. These mitotic defects are exacerbated by inhibition of ATR or WEE1 and therefore point to mitotic catastrophe as an underlying mechanism. Importantly, our data suggest that Cyclin E1 overexpression can be used to select patients for treatment with replication checkpoint inhibitors.
Conflict of interest statement
M.A.T.M.v.V. has acted on the Scientific Advisory Board of Repare Therapeutics, which is unrelated to this work. The other authors declare no conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

