Arabidopsis phenotyping reveals the importance of alcohol dehydrogenase and pyruvate decarboxylase for aerobic plant growth
- PMID: 33028901
- PMCID: PMC7542448
- DOI: 10.1038/s41598-020-73704-x
Arabidopsis phenotyping reveals the importance of alcohol dehydrogenase and pyruvate decarboxylase for aerobic plant growth
Abstract
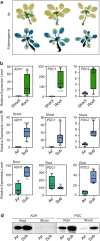
Alcohol dehydrogenase (ADH) and pyruvate decarboxylase (PDC) are key to the establishment of the fermentative metabolism in plants during oxygen shortage. Most of the evidence that both ADH and PDC are required for plant tolerance to hypoxia comes from experiments performed by limiting oxygen in the environment, such as by exposing plants to gaseous hypoxia or to waterlogging or submergence. However, recent experiments have shown that hypoxic niches might exist in plants grown in aerobic conditions. Here, we investigated the importance of ADH and PDC for plant growth and development under aerobic conditions, long-term waterlogging and short-term submergence. Data were collected after optimizing the software associated with a commercially-available phenotyping instrument, to circumvent problems in separation of plants and background pixels based on colour features, which is not applicable for low-oxygen stressed plants due to the low colour contrast of leaves with the brownish soil. The results showed that the growth penalty associated with the lack of functional ADH1 or both PDC1 and PDC2 is greater under aerobic conditions than in hypoxia, highlighting the importance of fermentative metabolism in plants grown under normal, aerobic conditions.
Conflict of interest statement
The authors declare the following competing interests: at the time of the study Tino Dornbusch was an employee of LemnaTec GmbH. The other authors have no competing interests as defined by Nature Research, or other interests that might be perceived to influence the results and/or discussion reported in this paper.
Figures







References
-
- Perata P, Alpi A. Plant responses to anaerobiosis. Plant Sci. 1993;93:1–17. doi: 10.1016/0168-9452(93)90029-Y. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

