S-adenosylmethionine: A metabolite critical to the regulation of autophagy
- PMID: 33030764
- PMCID: PMC7653241
- DOI: 10.1111/cpr.12891
S-adenosylmethionine: A metabolite critical to the regulation of autophagy
Abstract
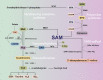
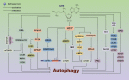
Autophagy is a mechanism that enables cells to maintain cellular homeostasis by removing damaged materials and mobilizing energy reserves in conditions of starvation. Although nutrient availability strongly impacts the process of autophagy, the specific metabolites that regulate autophagic responses have not yet been determined. Recent results indicate that S-adenosylmethionine (SAM) represents a critical inhibitor of methionine starvation-induced autophagy. SAM is primarily involved in four key metabolic pathways: transmethylation, transsulphuration, polyamine synthesis and 5'-deoxyadenosyl 5'-radical-mediated biochemical transformations. SAM is the sole methyl group donor involved in the methylation of DNA, RNA and histones, modulating the autophagic process by mediating epigenetic effects. Moreover, the metabolites of SAM, such as homocysteine, glutathione, decarboxylated SAM and spermidine, also exert important influences on the regulation of autophagy. From our perspective, nuclear-cytosolic SAM is a conserved metabolic inhibitor that connects cellular metabolic status and the regulation of autophagy. In the future, SAM might be a new target of autophagy regulators and be widely used in the treatment of various diseases.
© 2020 The Authors. Cell Proliferation Published by John Wiley & Sons Ltd.
Conflict of interest statement
No potential conflicts of interest were disclosed.
Figures
References
-
- Saha S, Panigrahi DP, Patil S, Bhutia SK. Autophagy in health and disease: a comprehensive review. Biomed Pharmacother. 2018;104:485‐495. - PubMed
-
- He CC, Klionsky DJ. Autophagy and neurodegeneration. ACS Chem Biol. 2006;1(4):211‐213. - PubMed
-
- Abdellatif M, Sedej S, Carmona‐Gutierrez D, Madeo F, Kroemer G. Autophagy in cardiovascular aging. Circ Res. 2018;123(7):803‐824. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- WJ2019Q053/Health Commission of Hubei Province Scientific Research Project
- 2012YQ160203/National Major Scientific Instruments and Equipment Development Projects
- 81302314/National Natural Science Foundation of China
- 81471781/National Natural Science Foundation of China
- 81903166/National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources

