Rapid Serological Assays and SARS-CoV-2 Real-Time Polymerase Chain Reaction Assays for the Detection of SARS-CoV-2: Comparative Study
- PMID: 33031048
- PMCID: PMC7641647
- DOI: 10.2196/19152
Rapid Serological Assays and SARS-CoV-2 Real-Time Polymerase Chain Reaction Assays for the Detection of SARS-CoV-2: Comparative Study
Abstract
Background: Real-time polymerase chain reaction (RT-PCR) testing for the identification of viral nucleic acid is the current standard for the diagnosis of SARS-CoV-2 infection, but technical issues limit its utilization for large-scale screening. Serological immunoglobulin M (IgM)/IgG testing has been proposed as a useful tool for detecting SARS-CoV-2 exposure.
Objective: The objective of our study was to compare the results of the rapid serological VivaDiag test for SARS-CoV-2-related IgM/IgG detection with those of the standard RT-PCR laboratory test for identifying SARS-CoV-2 nucleic acid.
Methods: We simultaneously performed both serological and molecular tests with a consecutive series of 191 symptomatic patients. The results provided by a new rapid serological colorimetric test for analyzing IgM/IgG expression were compared with those of RT-PCR testing for SARS-CoV-2 detection.
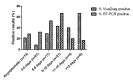
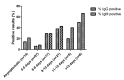
Results: Of the 191 subjects, 70 (36.6%) tested positive for SARS-CoV-2 based on RT-PCR results, while 34 (17.3%) tested positive based on serological IgM/IgG expression. Additionally, 13 (6.8%) subjects tested positive based on serological test results, but also tested negative based on RT-PCR results. The rapid serological test had a sensitivity of 30% and a specificity of 89% compared to the standard RT-PCR assay. Interestingly, the performance of both assays improved 8 days after symptom appearance. After 10 days had passed since symptom appearance, the predictive value of the rapid serological test was higher than that of the standard molecular assay (proportion of positive results: 40% vs 20%). Multivariate analysis showed that age >58 years (P<.01) and period of >15 days after symptom onset (P<.02) were significant and independent factors associated with serological test positivity.
Conclusions: The rapid serological test analyzed in this study seems limited in terms of usefulness when diagnosing SARS-CoV-2 infection. However, it may be useful for providing relevant information on people's immunoreaction to COVID-19 exposure.
Keywords: COVID-19; RT-PCR; SARS-CoV-2; serological test.
©Angelo Virgilio Paradiso, Simona De Summa, Daniela Loconsole, Vito Procacci, Anna Sallustio, Francesca Centrone, Nicola Silvestris, Vito Cafagna, Giuseppe De Palma, Antonio Tufaro, Vito Michele Garrisi, Maria Chironna. Originally published in the Journal of Medical Internet Research (http://www.jmir.org), 30.10.2020.
Conflict of interest statement
Conflicts of Interest: None declared.
Figures

References
-
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 Feb 15;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. https://linkinghub.elsevier.com/retrieve/pii/S0140-6736(20)30183-5 - DOI - PMC - PubMed
-
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020 Feb 20;382(8):727–733. doi: 10.1056/NEJMoa2001017. http://europepmc.org/abstract/MED/31978945 - DOI - PMC - PubMed
-
- Italy Coronavirus. Worldometer - real time world statistics. [2020-10-20]. https://www.worldometers.info/coronavirus/country/italy/
-
- Jin YH, Cai L, Cheng ZS, Cheng H, Deng T, Fan YP, Fang C, Huang D, Huang LQ, Huang Q, Han Y, Hu B, Hu F, Li BH, Li YR, Liang K, Lin LK, Luo LS, Ma J, Ma LL, Peng ZY, Pan YZ, Pan ZY, Ren XQ, Sun HM, Wang Y, Wang YY, Weng H, Wei CJ, Wu DF, Xia J, Xiong Y, Xu HB, Yao XM, Yuan YF, Ye TS, Zhang XC, Zhang YW, Zhang YG, Zhang HM, Zhao Y, Zhao MJ, Zi H, Zeng XT, Wang YY, Wang XH, for the Zhongnan Hospital of Wuhan University Novel Coronavirus Management and Research Team‚ Evidence-Based Medicine Chapter of China International Exchange and Promotive Association for Medical and Health Care (CPAM) A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version) Mil Med Res. 2020 Feb 06;7(1):4. doi: 10.1186/s40779-020-0233-6. https://mmrjournal.biomedcentral.com/articles/10.1186/s40779-020-0233-6 - DOI - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

